Abstract
Objective
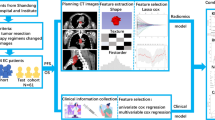
This study aimed to build radiomic feature-based machine learning models to predict pathological clinical response (pCR) of neoadjuvant chemoradiation therapy (nCRT) for esophageal squamous cell carcinoma (ESCC) patients.
Methods
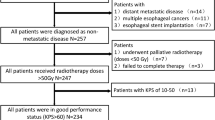
A total of 112 ESCC patients who underwent nCRT followed by surgical treatment from January 2008 to December 2018 were recruited. According to pCR status (no visible cancer cells in primary cancer lesion), patients were categorized into primary cancer lesion pCR (ppCR) group (N = 65) and non-ppCR group (N = 47). Patients were also categorized into total pCR (tpCR) group (N = 48) and non-tpCR group (N = 64) according to tpCR status (no visible cancer cells in primary cancer lesion or lymph nodes). Radiomic features of pretreatment CT images were extracted, feature selection was performed, machine learning models were trained to predict ppCR and tpCR, respectively.
Results
A total of 620 radiomic features were extracted. For ppCR prediction models, radiomic model had an area under the curve (AUC) of 0.817 (95% CI: 0.732–0.896) in the testing set; and the combination model that included rad-score and clinical features had a great predicting performance, with an AUC of 0.891 (95% CI: 0.823–0.950) in the testing set. For tpCR prediction models, radiomic model had an AUC of 0.713 (95% CI: 0.613–0.808) in the testing set; and the combination model also had a great predicting performance, with an AUC of 0.814 (95% CI: 0.728–0.881) in the testing set.
Conclusion
This study built machine learning models for predicting ppCR and tpCR of ESCC patients with favorable predicting performance respectively, which aided treatment plan optimization.
Clinical relevance statement
This study significantly improved the predictive value of machine learning models based on radiomic features to accurately predict response to therapy of esophageal squamous cell carcinoma patients after neoadjuvant chemoradiation therapy, providing guidance for further treatment.
Key Points
• Combination model that included rad-score and clinical features had a great predicting performance.
• Primary tumor pCR predicting models exhibit better predicting performance compared to corresponding total pCR predicting models.






Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the curve
- BMI:
-
Body mass index
- CT:
-
Computed tomography
- CTV:
-
Clinical target volume
- EC:
-
Esophageal cancer
- ESCC:
-
Esophageal squamous cell carcinoma
- GTV:
-
Gross tumor volume
- KPS:
-
Karnofsky performance status
- LOOCV:
-
Leave-one-out cross-validation
- MRI:
-
Magnetic resonance imaging
- nCRT:
-
Neoadjuvant chemoradiotherapy
- NPV:
-
Negative predictive value
- OAR:
-
Organs at risk
- OS:
-
Overall survival
- pCR:
-
Pathological complete response
- PD:
-
Program death
- PET:
-
Positron emission tomography
- PFS:
-
Progression-free survival
- ppCR:
-
Primary tumor lesion pCR
- PPV:
-
Positive predictive value
- PTV:
-
Plan target volume
- ROC:
-
Receiver operating characteristics
- TNR:
-
True negative rate
- tpCR:
-
Total pCR
- TPR:
-
True positive rate
- VOI:
-
Volumn of interest
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin 72: https://doi.org/10.3322/caac.21708
Eyck BM, van Lanschot JJB, Hulshof MCCM et al (2021) Ten-year outcome of neoadjuvant chemoradiotherapy plus surgery for esophageal cancer: the randomized controlled CROSS trial. J Clin Oncol 39:1995–2004. https://doi.org/10.1200/JCO.20.03614
Yang H, Liu H, Chen Y, et al (2018) Neoadjuvant chemoradiotherapy followed by surgery versus surgery alone for locally advanced squamous cell carcinoma of the esophagus (NEOCRTEC5010): a phase III multicenter, randomized, open-label clinical trial. J Clin Oncol 36: https://doi.org/10.1200/JCO.2018.79.1483
Donahue JM, Nichols FC, Li Z, et al (2009) Complete pathologic response after neoadjuvant chemoradiotherapy for esophageal cancer is associated with enhanced survival. Ann Thorac Surg 87: https://doi.org/10.1016/j.athoracsur.2008.11.001
Kelly RJ, Ajani JA, Kuzdzal J, et al (2021) Adjuvant nivolumab in resected esophageal or gastroesophageal junction cancer. N Engl J Med 384: https://doi.org/10.1056/nejmoa2032125
Dittrick GW, Weber JM, Shridhar R, et al (2012) Pathologic nonresponders after neoadjuvant chemoradiation for esophageal cancer demonstrate no survival benefit compared with patients treated with primary esophagectomy. Ann Surg Oncol 19: https://doi.org/10.1245/s10434-011-2078-4
Yang Z, He B, Zhuang X, et al (2019) CT-based radiomic signatures for prediction of pathologic complete response in esophageal squamous cell carcinoma after neoadjuvant chemoradiotherapy. J Radiat Res 60: https://doi.org/10.1093/jrr/rrz027
Qu J, Ma L, Lu Y, et al (2022) DCE-MRI radiomics nomogram can predict response to neoadjuvant chemotherapy in esophageal cancer. Discover Oncol 13: https://doi.org/10.1007/s12672-022-00464-7
van Rossum PSN, van Lier ALHMW, van Vulpen M, et al (2015) Diffusion-weighted magnetic resonance imaging for the prediction of pathologic response to neoadjuvant chemoradiotherapy in esophageal cancer. Radiother Oncol 115: https://doi.org/10.1016/j.radonc.2015.04.027
Tan S, Kligerman S, Chen W, et al (2013) Spatial-temporal [18F]FDG-PET features for predicting pathologic response of esophageal cancer to neoadjuvant chemoradiation therapy. Int J Radiat Oncol Biol Phys 85: https://doi.org/10.1016/j.ijrobp.2013.09.037
Zhu Y, Yao W, Xu BC, et al (2021) Predicting response to immunotherapy plus chemotherapy in patients with esophageal squamous cell carcinoma using non-invasive Radiomic biomarkers. BMC Cancer 21: https://doi.org/10.1186/s12885-021-08899-x
Zhang X, Gari A, Li M, et al (2022) Combining serum inflammation indexes at baseline and post treatment could predict pathological efficacy to anti PD 1 combined with neoadjuvant chemotherapy in esophageal squamous cell carcinoma. J Transl Med 20: https://doi.org/10.1186/s12967-022-03252-7
Toxopeus ELA, Nieboer D, Shapiro J, et al (2015) Nomogram for predicting pathologically complete response after neoadjuvant chemoradiotherapy for oesophageal cancer. Radiother Oncol 115: https://doi.org/10.1016/j.radonc.2015.04.028
van Rossum PSN, Fried DV, Zhang L, et al (2016) The incremental value of subjective and quantitative assessment of 18F-FDG PET for the prediction of pathologic complete response to preoperative chemoradiotherapy in esophageal cancer. J Nucl Med 57: https://doi.org/10.2967/jnumed.115.163766
Hu Y, Xie C, Yang H et al (2021) Computed tomography-based deep-learning prediction of neoadjuvant chemoradiotherapy treatment response in esophageal squamous cell carcinoma. Radiother Oncol 154:6–13. https://doi.org/10.1016/j.radonc.2020.09.014
Duong C, Greenawalt DM, Kowalczyk A et al (2007) Pretreatment gene expression profiles can be used to predict response to neoadjuvant chemoradiotherapy in esophageal cancer. Ann Surg Oncol 14:3602–3609. https://doi.org/10.1245/s10434-007-9550-1
Cheng H, Garrick DJ, Fernando RL (2017) Efficient strategies for leave-one-out cross validation for genomic best linear unbiased prediction. J Anim Sci Biotechnol 8:38. https://doi.org/10.1186/s40104-017-0164-6
Shao Z, Er MJ, Wang N (2016) An efficient leave-one-out cross-validation-based extreme learning machine (ELOO-ELM) with minimal user intervention. IEEE Trans Cybern 46:1939–1951. https://doi.org/10.1109/TCYB.2015.2458177
Jeong YS, Jeon M, Park JH et al (2021) Machine-learning-based approach to differential diagnosis in tuberculous and viral meningitis. Infect Chemother 53:53–62. https://doi.org/10.3947/ic.2020.0104
Cheng J, Dekkers JCM, Fernando RL (2021) Cross-validation of best linear unbiased predictions of breeding values using an efficient leave-one-out strategy. J Anim Breed Genet 138:519–527. https://doi.org/10.1111/jbg.12545
Nagata T, Noyori SS, Noguchi H et al (2021) Skin tear classification using machine learning from digital RGB image. J Tissue Viability 30:588–593. https://doi.org/10.1016/j.jtv.2021.01.004
Li M, Jin Y-M, Zhang Y-C et al (2021) Radiomics for predicting perineural invasion status in rectal cancer. World J Gastroenterol 27:5610–5621. https://doi.org/10.3748/wjg.v27.i33.5610
Xu W, Wu W, Zheng Y et al (2021) A computed tomography radiomics-based prediction model on interstitial lung disease in anti-MDA5-positive dermatomyositis. Front Med (Lausanne) 8:768052. https://doi.org/10.3389/fmed.2021.768052
Sun Y, Li C, Jin L et al (2020) Radiomics for lung adenocarcinoma manifesting as pure ground-glass nodules: invasive prediction. Eur Radiol 30:3650–3659. https://doi.org/10.1007/s00330-020-06776-y
Cui Y, Li Z, Xiang M et al (2022) Machine learning models predict overall survival and progression free survival of non-surgical esophageal cancer patients with chemoradiotherapy based on CT image radiomics signatures. Radiat Oncol 17:212. https://doi.org/10.1186/s13014-022-02186-0
Funding
This work was supported by the grants from the Zhejiang province public welfare funds (No. GF20H160009) and the Medical Science and Technology Project of Zhejiang Province (No. 2020382901, and 2021455891).
Author information
Authors and Affiliations
Contributions
WJ: data collection, statistical analysis, and writing and revising the manuscript. ZX, ZJ, LC, SW: statistical analysis and revising the manuscript. ZJ, SXJ: patient administration, and data collection. FJ, LQR: patient administration, and critical revision of the manuscript. JYL and CQX study design, statistical analysis, critical revision of the manuscript, and funds collection.
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication Yongling Ji.
Conflict of interest
The authors declare that they have no competing interests.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Not applicable.
Ethical approval
The study was approved by our hospital’s institutional review board.
Study subjects or cohorts overlap
No study subjects or cohorts have been previously reported.
Methodology
• retrospective
• observational
• performed at one institution
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, J., Zhu, X., Zeng, J. et al. Using clinical and radiomic feature–based machine learning models to predict pathological complete response in patients with esophageal squamous cell carcinoma receiving neoadjuvant chemoradiation. Eur Radiol 33, 8554–8563 (2023). https://doi.org/10.1007/s00330-023-09884-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-023-09884-7