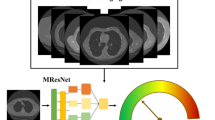
Abstract
Objective
To develop a deep learning–based artificial intelligence (AI) scheme for predicting the likelihood of the ground-glass nodule (GGN) detected on CT images being invasive adenocarcinoma (IA) and also compare the accuracy of this AI scheme with that of two radiologists.
Methods
First, we retrospectively collected 828 histopathologically confirmed GGNs of 644 patients from two centers. Among them, 209 GGNs are confirmed IA and 619 are non-IA, including 409 adenocarcinomas in situ and 210 minimally invasive adenocarcinomas. Second, we applied a series of pre-preprocessing techniques, such as image resampling, rescaling and cropping, and data augmentation, to process original CT images and generate new training and testing images. Third, we built an AI scheme based on a deep convolutional neural network by using a residual learning architecture and batch normalization technique. Finally, we conducted an observer study and compared the prediction performance of the AI scheme with that of two radiologists using an independent dataset with 102 GGNs.
Results
The new AI scheme yielded an area under the receiver operating characteristic curve (AUC) of 0.92 ± 0.03 in classifying between IA and non-IA GGNs, which is equivalent to the senior radiologist’s performance (AUC 0.92 ± 0.03) and higher than the score of the junior radiologist (AUC 0.90 ± 0.03). The Kappa value of two sets of subjective prediction scores generated by two radiologists is 0.6.
Conclusions
The study result demonstrates using an AI scheme to improve the performance in predicting IA, which can help improve the development of a more effective personalized cancer treatment paradigm.
Key Points
• The feasibility of using a deep learning method to predict the likelihood of the ground-glass nodule being invasive adenocarcinoma.
• Residual learning–based CNN model improves the performance in classifying between IA and non-IA nodules.
• Artificial intelligence (AI) scheme yields higher performance than radiologists in predicting invasive adenocarcinoma.





Similar content being viewed by others
Abbreviations
- AAH:
-
Atypical adenomatous hyperplasia
- ACC:
-
Accuracy
- AIS:
-
Adenocarcinoma in situ
- AUC:
-
Area under a ROC curve
- CADx:
-
Computer-aided diagnosis
- CNN:
-
Convolutional neural network
- CT:
-
Computed tomography
- DFS:
-
Disease-free survival
- FC:
-
Fully connected
- GGN:
-
Ground-glass nodules
- IAC:
-
Invasive adenocarcinoma
- MCC:
-
Matthews correlation coefficient
- MIA:
-
Minimally invasive adenocarcinoma
- NSCLC:
-
Non-small cell lung cancer
- QI:
-
Quantitative imaging
- ReLU:
-
Rectified linear unit
- ROC:
-
Receiver operating characteristic
References
Zappa C, Mousa SA (2016) Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res 5:288–300
Han L, Zhang P, Wang Y et al (2018) CT quantitative parameters to predict the invasiveness of lung pure ground-glass nodules (pGGNs). Clin Radiol 73:504.e1–504.e7
Zhang Y, Tang J, Xu J, Cheng J, Wu H (2017) Analysis of pulmonary pure ground-glass nodule in enhanced dual energy CTimaging for predicting invasive adenocarcinoma: comparing with conventional thin-section CT imaging. J Thorac Dis 9:4967–4978
Hu H, Wang Q, Tang H, Xiong L, Lin Q (2016) Multi-slice computed tomography characteristics of solitary pulmonary groundglass nodules: differences between malignant and benign. Thorac Cancer 7:80–87
MacMahon H, Naidich DP, Goo JM et al (2017) Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017. Radiology 284:228–243
Lee HY, La Choi Y, Lee KS et al (2014) Pure ground-glass opacity neoplastic lung nodules: histopathology, imaging, management. AJR Am J Roentgenol 202:224–233
Pedersen JH, Saghir Z, Wille MMW et al (2016) Ground-glass opacity lung nodules in the era of lung cancer CT screening: radiology, pathology, and clinical management. Oncology (Willston Park) 30:266–274
Travis WD, Brambilla E, Noguchi M et al (2011) International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 6:244–285
Fan L, Fang MJ, Li ZB et al (2019) Radiomics signature: a biomarker for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule. Eur Radiol 29:889–897
Li M, Wu N, Zhang L et al (2018) Solid component proportion is an important predictor of tumor invasiveness in clinical stage T1N0M0 (cT1N0M0) lung adenocarcinoma. Cancer Imaging 18:18
Wang S, Wang R, Zhang S et al (2018) 3D convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground- glass nodules with diameters ≤3 cm using HRCT. Quant Imaging Med Surg 8:491–499
Lee GD, Park CH, Park HS et al (2019) Lung adenocarcinoma invasiveness risk in pure ground-glass opacity lung nodules smaller than 2 cm. Thorac Cardiovasc Surg 67:321–328
Gong J, Liu J-Y, Sun X-W et al (2018) Computer-aided diagnosis of lung cancer: the effect of training data sets on classification accuracy of lung nodules. Phys Med Biol 63:035036
Gong J, Liu J, Jiang Y et al (2018) Fusion of quantitative imaging features and serum biomarkers to improve performance of computer-aided diagnosis scheme for lung cancer: A preliminary study. Med Phys 45:5472–5481
Gao F, Sun Y, Zhang G et al (2019) CTcharacterization of different pathological types of subcentimeter pulmonary ground-glass nodular lesions. Br J Radiol 92:20180204
Cohen JG, Goo JM, Yoo R et al (2016) Software performance in segmenting ground-glass and solid components of subsolid nodules in pulmonary adenocarcinomas. Eur Radiol 26:4465–4474
Wang XW, Chen WF, He WJ et al (2018) CT features differentiating pre- and minimally invasive from invasive adenocarcinoma appearing as mixed ground-glass nodules: mass is a potential imaging biomarker. Clin Radiol 73:549–554
Zhao W, Yang J, Sun Y et al (2018) 3D Deep Learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res 78:6881–6889
Yasaka K, Akai H, Kunimatsu A et al (2018) Liver fibrosis: deep convolutional neural network for staging by using gadoxetic acid–enhanced hepatobiliary phaseMRimages. Radiology 287:146–155
Shen D, Wu G, Suk H-I (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE 770-778
Srivastava N, Hinton GE, Krizhevsky A et al (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15:1929–1958
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv e-prints 1-11. Available via https://arxiv.org/abs/1502.03167. Accessed 11 Feb 2015
Metz CE (2006) Receiver operating characteristic analysis: a tool for the quantitative evaluation of observer performance and imaging systems. J Am Coll Radiol 3:413–422
Powers DM (2011) Evaluation: from precision, recall and Fmeasure to ROC, informedness, markedness and correlation. J Mach Learn Technol 2:37–63
Ben-David A (2008) About the relationship between ROC curves and Cohen’s kappa. Eng Appl Artif Intel 21:874–882
Son JY, Lee HY, Kim J-H et al (2016) Quantitative CT analysis of pulmonary ground-glass opacity nodules for distinguishing invasive adenocarcinoma from non-invasive or minimally invasive adenocarcinoma: the added value of using iodine mapping. Eur Radiol 26:43–54
Zhang YP, Heuvelmans MA, Zhang H et al (2018) Changes in quantitative CT image features of ground-glass nodules in differentiating invasive pulmonary adenocarcinoma from benign and in situ lesions: histopathological comparisons. Clin Radiol 73:504.e9–504.e16
Chae H-D, Park CM, Park SJ et al (2014) Computerized texture analysis of persistent part-solid ground-glass nodules: differentiation of preinvasive lesions from invasive pulmonary adenocarcinomas. Radiology 273:285–293
Funding
This work was partially funded by China Postdoctoral Science Foundation under Grant No. 2019M651372, the Shanghai Science and Technology Funds under Grant No. 13411950107, the Natural Science Foundation of Shanghai under Grant No. 14ZR1427900.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Weijun Peng.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gong, J., Liu, J., Hao, W. et al. A deep residual learning network for predicting lung adenocarcinoma manifesting as ground-glass nodule on CT images. Eur Radiol 30, 1847–1855 (2020). https://doi.org/10.1007/s00330-019-06533-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06533-w