Abstract
Purpose
To identify triple-negative (TN) breast cancer imaging biomarkers in comparison to other molecular subtypes using multiparametric MR imaging maps and whole-tumor histogram analysis.
Materials and methods
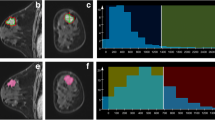
This retrospective study included 134 patients with invasive ductal carcinoma. Whole-tumor histogram-based texture features were extracted from a quantitative ADC map and DCE semi-quantitative maps (washin and washout). Univariate analysis using the Student’s t test or Mann–Whitney U test was performed to identify significant variables for differentiating TN cancer from other subtypes. The ROC curves were generated based on the significant variables identified from the univariate analysis. The AUC, sensitivity, and specificity for subtype differentiation were reported.
Results
The significant parameters on the univariate analysis achieved an AUC of 0.710 (95% confidence interval [CI] 0.562, 0.858) with a sensitivity of 63.6% and a specificity of 73.1% at the best cutoff point for differentiating TN cancers from Luminal A cancers. An AUC of 0.763 (95% CI 0.608, 0.917) with a sensitivity of 86.4% and a specificity of 72.2% was achieved for differentiating TN cancers from human epidermal growth factor receptor 2 (HER2) positive cancers. Also, an AUC of 0.683 (95% CI 0.556, 0.809) with a sensitivity of 54.5% and a specificity of 83.9% was achieved for differentiating TN cancers from non-TN cancers. There was no significant feature on the univariate analysis for TN cancers versus Luminal B cancers.
Conclusions
Whole-tumor histogram-based imaging features derived from ADC, along with washin and washout maps, provide a non-invasive analytical approach for discriminating TN cancers from other subtypes.
Key Points
• Whole-tumor histogram-based features on MR multiparametric maps can help to assess biological characterization of breast cancer.
• Histogram-based texture analysis may predict the molecular subtypes of breast cancer.
• Combined DWI and DCE evaluation helps to identify triple-negative breast cancer.




Similar content being viewed by others
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- AUC:
-
Area under the curve
- DCE:
-
Dynamic contrast-enhanced imaging
- DWI:
-
Diffusion-weighted imaging
- ER:
-
Estrogen receptor
- HER2:
-
Human epidermal growth factor receptor 2
- IDC:
-
Invasive ductal carcinoma
- IHC:
-
Immunohistochemical
- PR:
-
Progesterone receptor
- SD:
-
Standard deviation
- SI:
-
Signal intensity
- TN:
-
Triple-negative
References
Wu JM, Fackler MJ, Halushka MK et al (2008) Heterogeneity of breast cancer metastases: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res 14:1938–1946
Goldhirsch A, Wood WC, Coates AS et al (2011) Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann Oncol 22:1736–1747
Dent R, Trudeau M, Pritchard KI et al (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13:4429–4434
Liedtke C, Mazouni C, Hess KR et al (2008) Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 26:1275–1281
Cleator S, Heller W, Coombes RC (2007) Triple-negative breast cancer: therapeutic options. Lancet Oncol 8:235–244
Loo CE, Straver ME, Rodenhuis S et al (2011) Magnetic resonance imaging response monitoring of breast cancer during neoadjuvant chemotherapy: relevance of breast cancer subtype. J Clin Oncol 29:660–666
Youk JH, Son EJ, Chung J, Kim JA, Kim EK (2012) Triple-negative invasive breast cancer on dynamic contrast-enhanced and diffusion-weighted MR imaging: comparison with other breast cancer subtypes. Eur Radiol 22:1724–1734
Kato F, Kudo K, Yamashita H et al (2016) Differences in morphological features and minimum apparent diffusion coefficient values among breast cancer subtypes using 3-tesla MRI. Eur J Radiol 85:96–102
Chen JH, Agrawal G, Feig B et al (2007) Triple-negative breast cancer: MRI features in 29 patients. Ann Oncol 18:2042–2043
Koo HR, Cho N, Song IC et al (2012) Correlation of perfusion parameters on dynamic contrast-enhanced MRI with prognostic factors and subtypes of breast cancers. J Magn Reson Imaging 36:145–151
Uematsu T, Kasami M, Yuen S (2009) Triple-negative breast cancer: correlation between MR imaging and pathologic findings. Radiology 250:638–647
Zhang B, Chang K, Ramkissoon S et al (2017) Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol 19:109–117
Gibbs P, Turnbull LW (2003) Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med 50:92–98
Sinha S, Lucas-Quesada FA, DeBruhl ND et al (1997) Multifeature analysis of Gd-enhanced MR images of breast lesions. J Magn Reson Imaging 7:1016–1026
Agner SC, Rosen MA, Englander S et al (2014) Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: a feasibility study. Radiology 272:91–99
Chaudhury B, Zhou M, Goldgof DB et al (2015) Heterogeneity in intratumoral regions with rapid gadolinium washout correlates with estrogen receptor status and nodal metastasis. J Magn Reson Imaging 42:1421–1430
Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI (2014) Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 273:365–372
Waugh SA, Purdie CA, Jordan LB et al (2016) Magnetic resonance imaging texture analysis classification of primary breast cancer. Eur Radiol 26:322–330
Kim EJ, Kim SH, Park GE et al (2015) Histogram analysis of apparent diffusion coefficient at 3.0t: correlation with prognostic factors and subtypes of invasive ductal carcinoma. J Magn Reson Imaging 42:1666–1678
Choi Y, Kim SH, Youn IK, Kang BJ, Park WC, Lee A (2017) Rim sign and histogram analysis of apparent diffusion coefficient values on diffusion-weighted MRI in triple-negative breast cancer: comparison with ER-positive subtype. PLoS One. https://doi.org/10.1371/journal.phone.0177903
Harms SE (1999) Technical report of the international working group on breast MRI. J Magn Reson Imaging 10:979
Grady L (2006) Random walks for image segmentation. IEEE Trans Pattern Anal Mach Intell 28:1768–1783
Koh DM, Collins DJ (2007) Diffusion-weighted MRI in the body: applications and challenges in oncology. AJR Am J Roentgenol 188:1622–1635
Martincich L, Deantoni V, Bertotto I et al (2012) Correlations between diffusion-weighted imaging and breast cancer biomarkers. Eur Radiol 22:1519–1528
Liu S, Ren R, Chen Z et al (2015) Diffusion-weighted imaging in assessing pathological response of tumor in breast cancer subtype to neoadjuvant chemotherapy. J Magn Reson Imaging 42:779–787
Padhani AR, Liu G, Koh DM et al (2009) Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia 11:102–125
Park SH, Moon WK, Cho N et al (2010) Diffusion-weighted MR imaging: pretreatment prediction of response to neoadjuvant chemotherapy in patients with breast cancer. Radiology 257:56–63
Rahbar H, Partridge SC, Demartini WB et al (2012) In vivo assessment of ductal carcinoma in situ grade: a model incorporating dynamic contrast-enhanced and diffusion-weighted breast MR imaging parameters. Radiology 263:374–382
Narisada H, Aoki T, Sasaguri T et al (2006) Correlation between numeric gadolinium-enhanced dynamic MRI ratios and prognostic factors and histologic type of breast carcinoma. AJR Am J Roentgenol 187:297–306
Schmitz AM, Loo CE, Wesseling J, Pijnappel RM, Gilhuijs KG (2014) Association between rim enhancement of breast cancer on dynamic contrast-enhanced MRI and patient outcome: impact of subtype. Breast Cancer Res Treat 148:541–551
Blaschke E, Abe H (2015) MRI phenotype of breast cancer: kinetic assessment for molecular subtypes. J Magn Reson Imaging 42:920–924
Kuhl CK, Mielcareck P, Klaschik S et al (1999) Dynamic breast MR imaging: are signal intensity time course data useful for differential diagnosis of enhancing lesions? Radiology 211:101–110
Leong LC, Gombos EC, Jagadeesan J, Fook-Chong SM (2015) MRI kinetics with volumetric analysis in correlation with hormonal receptor subtypes and histologic grade of invasive breast cancers. AJR Am J Roentgenol 204:W348–W356
Brown LF, Berse B, Jackman RW et al (1995) Expression of vascular permeability factor (vascular endothelial growth factor) and its receptors in breast cancer. Hum Pathol 26:86–91
Kumar R, Yarmand-Bagheri R (2001) The role of HER2 in angiogenesis. Semin Oncol 28:27–32
Esteva FJ, Hortobagyi GN (2004) Prognostic molecular markers in early breast cancer. Breast Cancer Res 6:109–118
Lambregts DM, Beets GL, Maas M et al (2011) Tumour ADC measurements in rectal cancer: effect of ROI methods on ADC values and interobserver variability. Eur Radiol 21:2567–2574
Stoutjesdijk MJ, Futterer JJ, Boetes C, van Die LE, Jager G, Barentsz JO (2005) Variability in the description of morphologic and contrast enhancement characteristics of breast lesions on magnetic resonance imaging. Invest Radiol 40:355–362
Robson MD, Anderson AW, Gore JC (1997) Diffusion-weighted multiple shot echo planar imaging of humans without navigation. Magn Reson Med 38:82–88
Sutton EJ, Dashevsky BZ, Oh JH et al (2016) Breast cancer molecular subtype classifier that incorporates MRI features. J Magn Reson Imaging 44:122–129
Kim JH, Ko ES, Lim Y et al (2017) Breast cancer heterogeneity: MR imaging texture analysis and survival outcomes. Radiology 282:665–675
Funding
This study has received funding by the National Natural Science Foundation of China (61731008).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Weijun Peng.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
This study is retrospective study and does not require informed consent.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Electronic supplementary material
ESM 1
(DOCX 80 kb)
Rights and permissions
About this article
Cite this article
Xie, T., Zhao, Q., Fu, C. et al. Differentiation of triple-negative breast cancer from other subtypes through whole-tumor histogram analysis on multiparametric MR imaging. Eur Radiol 29, 2535–2544 (2019). https://doi.org/10.1007/s00330-018-5804-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-018-5804-5