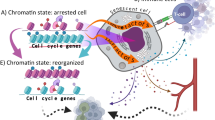
Abstract
Cellular senescence can induce dual effects (promotion or inhibition) on cancer progression. While immune cells naturally respond and migrate toward various chemotactic sources from the tumor mass, various factors including senescent tumor cells (STCs) in the tumor microenvironment may affect this chemotactic movement. In this work, we investigate the mutual interactions between the tumor cells and the immune cells that either inhibit or facilitate tumor growth by developing a mathematical model that consists of taxis–reaction–diffusion equations and receptor kinetics for the key players in the interaction network. We apply a mathematical model to a transwell Boyden chamber invasion assay used in the experiments to illustrate that STCs can play a pivotal role in negating immune attack through tight regulation of intra- and extra-cellular signaling molecules. In particular, we show that senescent tumor cells in cell cycle arrest can block intratumoral infiltration of CD8+ T cells by secreting a high level of CXCL12, which leads to significant reduction its receptors, CXCR4, on T cells, and thus impaired chemotaxis. The predictions of nonlinear responses to CXCL12 were in good agreement with experimental data. We tested several hypotheses on immune-tumor interactions under various biochemical conditions in the tumor microenvironment and developed new concepts for anti-tumor strategies targeting senescence induced immune impairment.
















Similar content being viewed by others
References
Akakura N, Horiuchi MK, Suzuki A, Wang J, Chen J, Niizeki H, Kawamura K, Hosokawa M, Asaka M (2001) Constitutive expression of hypoxia-inducible factor-1alpha renders pancreatic cancer cells resistant to apoptosis induced by hypoxia and nutrient deprivation. Cancer Res 61(17):6548–54
Aktas O, Ozturk A, Erman B, Erus S, Tanju S, Dilege S (2018) Role of natural killer cells in lung cancer. J Cancer Res Clin Oncol 144(6):997–1003
Aspirin AP, DelosReyes VAA, Kim Y (2021) Polytherapeutic strategies with oncolytic virus-bortezomib and adjuvant NK cells in cancer treatment. J R Soc Interface 18(174):20200669
Balkwill F (2012) The chemokine system and cancer. J Pathol 226(2):148–57
Batlle E, Clevers H (2017) Cancer stem cells revisited. Nat Med 23(10):1124–1134
Brahimi-Horn M, Chiche J, Pouyssegur J (2007) Hypoxia and cancer. J Mol Med (Berl) 85(12):1301–7
Brahmer J, Tykodi S, Chow L, Hwu W, Topalian S, Hwu P, Drake C, Camacho L, Kauh J, Odunsi K, Pitot H, Hamid O, Bhatia S, Martins R, Eaton K, Chen S, Salay T, Alaparthy S, Grosso J, Korman A, Parker S, Agrawal S, Goldberg SM, Pardoll D, Gupta A, Wigginton J (2012) Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 366(26):2455–65
Braig M, Lee S, Loddenkemper C, Rudolph C, Peters A, Schlegelberger B et al (2005) Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436(7051):660–5
Brown D (1999) Dependence of neurones on astrocytes in a coculture system renders neurones sensitive to transforming growth factor beta1-induced glutamate toxicity. J Neurochem 72(3):943–53
Bruni D, Angell H, Galon J (2020) The immune contexture and immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer 20(11):662–680
Campisi J (2013) Aging, cellular senescence, and cancer. Annu Rev Physiol 75:685–705
Cavnar S, Ray P, Moudgil P, Chang S, Luker K, Linderman J, Takayama S, Luker G (2014) Microfluidic source-sink model reveals effects of biophysically distinct CXCL12 isoforms in breast cancer chemotaxis. Integr Biol (Camb) 6(5):564–76
Chen X, Beutler J, McCloud T, Loehfelm A, Yang L, Dong H, Chertov O, Salcedo R, Oppenheim J, Howard O (2003) Tannic acid is an inhibitor of CXCL12 (SDF-1alpha)/CXCR4 with antiangiogenic activity. Clin Cancer Res 9(8):3115–23
Choi Y, Kim Y, Oh S, Suh K, Kim Y, Lee G, Yoon J, Park S, Lee Y, Park Y, Kim H, Park S, Kim J, Park T (2021) Senescent tumor cells build a cytokine shield in colorectal cancer. Adv Sci 8(4):2002497
Clercq ED (2015) AMD3100/CXCR4 inhibitor. Front Immunol 6:276
Collado M, Blasco M, Serrano M (2007) Cellular senescence in cancer and aging. Cell 130(2):223–33
Dallon JC, Othmer HG (1997) A discrete cell model with adaptive signalling for aggregation of dictyostelium discoideum. Philos Trans R Soc Lond B352:391–417
de Pillis L, Radunskaya A, Wiseman C (2005) A validated mathematical model of cell-mediated immune response to tumor growth. Cancer Res 65(17):7950
Decaillot F, Kazmi M, Lin Y, Raysaha S, Sakmar T, Sachdev P (2011) CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration. J Biol Chem 286(37):32188–32197
DiPersio J, Uy G, Yasothan U, Kirkpatrick P (2009) Plerixafor. Nat Rev Drug Discov 8(2):105–6
Drenckhan A, Kurschat N, Dohrmann T, Raabe N, Koenig A, Reichelt U, Kaifi J, Izbicki J, Gros S (2013) Effective inhibition of metastases and primary tumor growth with CTCE-9908 in esophageal cancer. J Surg Res 182(2):250–6
Duanghathaipornsuk S, Farrell E, Alba-Rubio A, Zelenay P, Kim D (2021) Detection technologies for reactive oxygen species: fluorescence and electrochemical methods and their applications. Biosensors (Basel) 11(2):30
Eftimie R, Barelle C (2021) Mathematical investigation of innate immune responses to lung cancer: the role of macrophages with mixed phenotypes. J Theor Biol 524:110,739
Esencay M, Newcomb E, Zagzag D (2010) HGF upregulates CXCR4 expression in gliomas via NF-kappaB: implications for glioma cell migration. J Neurooncol 99(1):33–40
Espejo E, Stevens A, Velazquez J (2009) Simultaneous finite time blow-up in a two-species model for chemotaxis. Analysis 29:317–338
Espejo E, Stevens A, Velazquez J (2010) A note on non-simultaneous blow-up for a drift-diffusion model. Differ Integr Equ 23(5/6):451–462
Espejo E, Stevens A, Suzuki T (2014) Simultaneous blowup and mass separation during collapse in an interacting systems of chemotactic species. Differ Integr Equ 25(3/4):251–288
Fahham D, Weiss I, Abraham M, Beider K, Hanna W, Shlomai Z, Eizenberg O, Zamir G, Izhar U, Shapira O, Peled A, Wald O (2012) In vitro and in vivo therapeutic efficacy of CXCR4 antagonist BKT140 against human non-small cell lung cancer. J Thorac Cardiovasc Surg 144(5):1167–1175
Fonkoua LK, Sirpilla O, Sakemura R, Siegler E, Kenderian S (2022) CAR T cell therapy and the tumor microenvironment: current challenges and opportunities. Mol Ther Oncolytics 25:69–77
Frascoli F, Kim P, Hughes B, Landman K (2014) A dynamical model of tumour immunotherapy. Math Biosci 253:50–62
Friedman A, Huang C, Yong J (1995) Effective permeability of the boundary of a domain. Commun. Partial Differ Equ 20(1 &2):59–102
Friedman A, Tian J, Fulci G, Chiocca E, Wang J (2006) Glioma virotherapy: effects of innate immune suppression and increased viral replication capacity. Cancer Res 66(4):2314–9
Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc P, Trajanoski Z, Fridman W, Pages F (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313(5795):1960–4
Ghosh M, Makena P, Gorantla V, Sinclair S, Waters C (2012) Cxcr4 regulates migration of lung alveolar epithelial cells through activation of rac1 and matrix metalloproteinase-2. Am J Physiol Lung Cell Mol Physiol 302(9):L846-56
Gil M, Seshadri M, Komorowski M, Abrams S, Kozbor D (2013) Targeting CXCL12/CXCR4 signaling with oncolytic virotherapy disrupts tumor vasculature and inhibits breast cancer metastases. Proc Natl Acad Sci USA 110(14):E1291-300
Golby S, Chinyama C, Spencer J (2002) Proliferation of T-cell subsets that contact tumour cells in colorectal cancer. Clin Exp Immunol 127(1):85–91
Gonzalez-Meljem J, Apps J, Fraser H, Martinez-Barbera J (2018) Paracrine roles of cellular senescence in promoting tumourigenesis. Br J Cancer 118(10):1283–1288
Guo F, Wang Y, Liu J, Mok S, Xue F, Zhang W (2016) CXCL12/CXCR4: a symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene 35(7):816–26
Hanes M, Salanga C, Chowdry A, Comerford I, Mccoll S, Kufareva I, Handel T (2015) Dual targeting of the chemokine receptors cxcr4 and ackr3 with novel engineered chemokines. J Biol Chem 290(37):22385–22397
Haugstetter A, Loddenkemper C, Lenze D, Grone J, Standfuss C, Petersen I, Dorken B, Schmitt C (2010) Cellular senescence predicts treatment outcome in metastasised colorectal cancer. Br J Cancer 103(4):505–9
Hendrix C, Flexner C, MacFarland R, Giandomenico C, Fuchs E, Redpath E, Bridger G, Henson G (2000) Pharmacokinetics and safety of AMD-3100, a novel antagonist of the CXCR-4 chemokine receptor, in human volunteers. Antimicrob Agents Chemother 44(6):1667–73
Hernandez-Segura A, Nehme J, Demaria M (2018) Hallmarks of cellular senescence. Trends Cell Biol 28(6):436–453
Hlavata L, Aguilaniu H, Pichova A, Nystrom T (2003) The oncogenic RAS2(val19) mutation locks respiration, independently of PKA, in a mode prone to generate ROS. EMBO J 22(13):3337–45
Hoffmann F, Muller W, Schutz D, Penfold M, Wong Y, Schulz S, Stumm R (2012) Rapid uptake and degradation of CXCL12 depend on CXCR7 carboxyl-terminal serine/threonine residues. J Biol Chem 287(34):28362–28377
Hoffman F, Gavaghan D, Osborne J, Barrett I, You T, Ghadially H, Sainson R, Wilkinson R, Byrne H (2018) A mathematical model of antibody-dependent cellular cytotoxicity (adcc). J Theor Biol 436:39–50
Huang Y, Hsiao Y, Chen Y, Wei Y, Lai T, Tang C (2007) Stromal cell-derived factor-1 enhances motility and integrin up-regulation through CXCR4, ERK and NF-kappaB-dependent pathway in human lung cancer cells. Biochem Pharmacol 74(12):1702–12
Jeon NL, Baskaran H, Dertinger SK, Whitesides GM, de Water LV, Toner M (2002) Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat Biotechnol 20(8):826–30
Kamijo T, Zindy F, Roussel M, Quelle D, Downing J, Ashmun R, Grosveld G, Sherr C (1997) Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91(5):649–59
Kato I, Niwa A, Heike T, Fujino H, Saito M, Umeda K, Hiramatsu H, Ito M, Morita M, Nishinaka Y, Adachi S, Ishikawa F, Nakahata T (2011) Identification of hepatic niche harboring human acute lymphoblastic leukemic cells via the SDF-1/CXCR4 axis. PLoS ONE 6(11):e27,042
Kim Y (2013) Regulation of cell proliferation and migration in glioblastoma: new therapeutic approach. Front Oncol 3:53
Kim Y, Friedman A (2010) Interaction of tumor with its microenvironment: a mathematical model. Bull Math Biol 72(5):1029–1068
Kim Y, Othmer H (2013) A hybrid model of tumor-stromal interactions in breast cancer. Bull Math Biol 75:1304–1350
Kim Y, Roh S (2013) A hybrid model for cell proliferation and migration in glioblastoma. Discrete Continu Dyn Syst B 18(4):969–1015
Kim Y, Othmer H (2015) Hybrid models of cell and tissue dynamics in tumor growth. Math Bios Eng 12(6):1141–1156
Kim Y, Park T (2019) Cellular senescence in cancer. BMB Rep 52(1):42–46
Kim Y, Stolarska M, Othmer H (2007) A hybrid model for tumor spheroid growth in vitro I: theoretical development and early results. Math Models Methods Appl Sci 17:1773–1798
Kim Y, Lawler S, Nowicki M, Chiocca E, Friedman A (2009) A mathematical model of brain tumor: pattern formation of glioma cells outside the tumor spheroid core. J Theor Biol 260:359–371
Kim Y, Wallace J, Li F, Ostrowski M, Friedman A (2010) Transformed epithelial cells and fibroblasts/myofibroblasts interaction in breast tumor: a mathematical model and experiments. J Math Biol 61(3):401–421
Kim Y, Stolarska M, Othmer H (2011) The role of the microenvironment in tumor growth and invasion. Prog Biophys Mol Biol 106:353–379
Kim Y, Jung T, Jung S, Jang W, Moon K, Kim I, Lee M, Lee J (2012) Tumour-infiltrating T-cell subpopulations in glioblastomas. Br J Neurosurg 26(1):21–7
Kim Y, Powathil G, Kang H, Trucu D, Kim H, Lawler S, Chaplain M (2015) Strategies of eradicating glioma cells: a multi-scale mathematical model with miR-451-AMPK-mTOR control. PLoS ONE 10(1):e0114370
Kim Y, Choi Y, Lee J, Soh E, Kim J, Park T (2017a) Senescent tumor cells lead the collective invasion in thyroid cancer. Nat Commun 8:15,208
Kim Y, Jeon H, Othmer H (2017b) The role of the tumor microenvironment in glioblastoma: a mathematical model. IEEE Trans Biomed Eng 64(3):519–527
Kim Y, Kang H, Powathil G, Kim H, Trucu D, Lee W, Lawler S, Chaplain M (2018a) Role of extracellular matrix and microenvironment in regulation of tumor growth and lar-mediated invasion in glioblastoma. PLoS ONE 13(10):e0204,865
Kim Y, Yoo J, Lee T, Liu J, Yu J, Caligiuri M, Kaur B, Friedman A (2018b) Complex role of NK cells in regulation of oncolytic virus-bortezomib therapy. Proc Natl Acad Sci USA 115(19):4927–4932
Kim Y, Lee D, Lee J, Lee S, Lawler S (2019a) Role of tumor-associated neutrophils in regulation of tumor growth in lung cancer development: a mathematical model. PLoS ONE 14(1):e0211041
Kim Y, Lee J, Lee D, Othmer H (2019b) Synergistic effects of bortezomib-OV therapy and anti-invasive strategies in glioblastoma: a mathematical model. Cancers 11:215
Kim Y, Lee D, Lawler S (2020) Collective invasion of glioma cells through OCT1 signalling and interaction with reactive astrocytes after surgery. Philos Trans R Soc B 375:20190390
Koka S, Vance J, Maze G (1995) Bone growth factors: potential for use as an osseointegration enhancement technique (oet). J West Soc Periodontol Periodontal Abstr 43(3):97–104
Kuhne M, Mulvey T, Belanger B, Chen S, Pan C, Chong C, Cao F, Niekro W, Kempe T, Henning K, Cohen L, Korman A, Cardarelli P (2013) BMS-936564/MDX-1338: a fully human anti-CXCR4 antibody induces apoptosis in vitro and shows antitumor activity in vivo in hematologic malignancies. Clin Cancer Res 19(2):357–66
Larson B, Rieger L, Instruments B (2015) Resources-application notes combining kinetic ligand binding and 3d tumor invasion technologies to assess drug residence time and anti-metastatic effects of CXCR4 inhibitors
Lee J, Lee D, Lawler S, Kim Y (2021) Role of neutrophil extracellular traps in regulation of lung cancer invasion and metastasis: structural insights from a computational model. PLoS Comput Biol 17(2):e1008257
Lefort S, Thuleau A, Kieffer Y, Sirven P, Bieche I, Marangoni E, Vincent-Salomon A, Mechta-Grigoriou F (2017) CXCR4 inhibitors could benefit to HER2 but not to triple-negative breast cancer patients. Oncogene 36(9):1211–1222
Leone V, D’Angelo D, Rubio I, de Freitas P, Federico A, Colamaio M, Pallante P, Medeiros-Neto G, Fusco A (2011) MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1alpha. J Clin Endocrinol Metab 96(9):E1388-98
Li X, Gruosso T, Zuo D, Omeroglu A, Meterissian S, Guiot M, Salazar A, Park M, Levine H (2019) Infiltration of CD8+ T cells into tumor cell clusters in triple-negative breast cancer. Proc Natl Acad Sci USA 116(9):3678–3687
Liang S, Hoskins M, Khanna P, Kunz R, Dong C (2008) Effects of the tumor-leukocyte microenvironment on melanoma-neutrophil adhesion to the endothelium in a shear flow. Cell Mol Bioeng 1(2–3):189–200
Liao W, Overman M, Boutin A, Shang X, Zhao D, Dey P, Li J, Wang G, Lan Z, Li J, Tang M, Jiang S, Ma X, Chen P, Katkhuda R, Korphaisarn K, Chakravarti D, Chang A, Spring D, Chang Q, Zhang J, Maru D, Maeda D, Zebala J, Kopetz S, Wang Y, DePinho R (2019) KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell 35(4):559–572
Lodish H, Berk A, Zipursky S, Matsudaira P, Baltimore D, Darnell J (2000) Molecular cell biology, 4th edn. W. H. Freeman, New York
Lukaszewicz-Zajac M, Mroczko B (2016) The serum concentrations of chemokine CXCL12 and its specific receptor CXCR4 in patients with esophageal cancer. Dis Markers 7963:895
Macheret M, Halazonetis T (2015) DNA replication stress as a hallmark of cancer. Annu Rev Pathol 10:425–48
Mackay C (1996) Chemokine receptors and T cell chemotaxis. J Exp Med 184(3):799–802
Mahlbacher G, Curtis L, Lowengrub J, Frieboes H (2018) Mathematical modeling of tumor-associated macrophage interactions with the cancer microenvironment. J Immunother Cancer 6(1):10
Mantovani A (2004) Chemokines in neoplastic progression. Semin Cancer Biol 14(3):147–8
Mantovani A, Allavena P, Sozzani S, Vecchi A, Locati M, Sica A (2004) Chemokines in neoplastic progression. Semin Cancer Biol 14(3):155–60
Marino S, Hogue I, Ray C, Kirschner D (2008) A methodology for performing global uncertainty and sensitivity analysis in systems biology. J Theor Biol 254(1):178–96
Mauldin I, Jo J, Wages N, Yogendran L, Mahmutovic A, Young S, Lopes M, Erickson L, Fadul C (2021) Proliferating CD8+ T cell infiltrates are associated with improved survival in glioblastoma. Cells 10(12):3378
Mellman I, Coukos G, Dranoff G (2011) Cancer immunotherapy comes of age. Nature 480(7378):480–9
Mhessn R, Abd-Alredha L, Al-Rubaie R, Aziz A (2011) Preparation of tannin based hydrogel for biological application. J Chem 8:763,295
Michaloglou C, Vredeveld L, Soengas M, Denoyelle C, Kuilman T, van der Horst C, Majoor D, Shay J, Mooi W, Peeper D (2005) BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436(7051):720–4
Milanovic M, Fan D, Belenki D, Dabritz J, Zhao Z, Yu Y, Dorr J, Dimitrova L, Lenze D, Barbosa IM, Mendoza-Parra M, Kanashova T, Metzner M, Pardon K, Reimann M, Trumpp A, Dorken B, Zuber J, Gronemeyer H, Hummel M, Dittmar G, Lee S, Schmitt C (2018) Senescence-associated reprogramming promotes cancer stemness. Nature 553(7686):96–100
Millis S, Ikeda S, Reddy S, Gatalica Z, Kurzrock R (2016) Landscape of phosphatidylinositol-3-kinase pathway alterations across diverse solid tumors. JAMA Oncol 2(12):1565–1573
Mlecnik B, Bindea G, Angell H, Maby P, Angelova M, Tougeron D, Church S, Lafontaine L, Fischer M, Fredriksen T, Sasso M, Bilocq A, Kirilovsky A, Obenauf A, Hamieh M, Berger A, Bruneval P, Tuech J, Sabourin J, Pessot FL, Mauillon J, Rafii A, Laurent-Puig P, Speicher M, Trajanoski Z, Michel P, Sesboue R, Frebourg T, Pages F, Valge-Archer V, Latouche J, Galon J (2016) Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 44(3):698–711
Nakano O, Sato M, Naito Y, Suzuki K, Orikasa S, Aizawa M, Suzuki Y, Shintaku I, Nagura H, Ohtani H (2001) Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: clinicopathologic demonstration of antitumor immunity. Cancer Res 61(13):5132–6
Nishihara H, Soldati S, Mossu A, Rosito M, Rudolph H, Muller W, Latorre D, Sallusto F, Sospedra M, Martin R, Ishikawa H, Tenenbaum T, Schroten H, Gosselet F, Engelhardt B (2020) Human CD4+ T cell subsets differ in their abilities to cross endothelial and epithelial brain barriers in vitro. Fluids Barriers CNS 17(1):3
O’Donoghue J, Bardies M, Wheldon T (1995) Relationships between tumor size and curability for uniformly targeted therapy with beta-emitting radionuclides. J Nucl Med 36(10):1902–9
Orlikowsky T, Neunhoeffer F, Goelz R, Eichner M, Henkel C, Zwirner M, Poets C (2004) Evaluation of IL-8-concentrations in plasma and lysed EDTA-blood in healthy neonates and those with suspected early onset bacterial infection. Pediatr Res 56(5):804–9
Othmer H, Stevens A (1997) Aggregation, blowup, and collapse: the abc’s of taxis in reinforced random walks. SIAM J Appl Math 57(4):1044–1081
Pages F, Mlecnik B, Marliot F, Bindea G, Ou F, Bifulco C et al (2018) International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 391(10135):2128–2139
Park S (2017) Survive or thrive: tradeoff strategy for cellular senescence. Exp Mol Med 49(6):e342
Park J, Wysocki R, Amoozgar Z, Maiorino L, Fein M, Jorns J, Schott A, Kinugasa-Katayama Y, Lee Y, Won N, Nakasone E, Hearn S, Kuttner V, Qiu J, Almeida A, Perurena N, Kessenbrock K, Goldberg M, Egeblad M (2016) Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci Transl Med 8(361):361ra138
Park S, Choi Y, Kim J, Kim H, Park T (2021a) Senescent tumor cells: an overlooked adversary in the battle against cancer. Exp Mol Med 53(12):1834–1841
Park J, Ryu S, Kim B, Cho H, Park C, Choi H, Jang E, Yang E, Hwang J, Woo S, Lee J, Park J, Choi K, Kwon Y, Lee C, Park J, Cho S, Lee Y, Lee S, Han J, Cho K, Kim M, Hwang D, Lee Y, Park S (2021b) Disruption of nucleocytoplasmic trafficking as a cellular senescence driver. Exp Mol Med 53(6):1092–1108
Peled A, Wald O, Burger J (2012) Development of novel CXCR4-based therapeutics. Expert Opin Investig Drugs 21(3):341–53
Pettet GJ, Byrne HM, McElwain DLS, Norbury J (1996) A model of wound healing angiogenesis in soft-tissue. Math Bios 136:35–64
Portella L, Bello A, Scala S (2021) CXCL12 signaling in the tumor microenvironment. Adv Exp Med Biol 1302:51–70
Prior I, Lewis P, Mattos C (2012) A comprehensive survey of RAS mutations in cancer. Cancer Res 72(10):2457–67
Rascle M, Ziti C (1995) Finite time blow up in some models of chemotaxis. J Math Biol 33:388–414
Redl H, Schlag G, Bahrami S, Dinges H, Schade U, Ceska M (1991a) Markers of endotoxin related leukocyte activation and injury mechanisms. Prog Clin Biol Res 367:83–100
Redl H, Schlag G, Bahrami S, Schade U, Ceska M, Stutz P (1991b) Plasma neutrophil-activating peptide-1/interleukin-8 and neutrophil elastase in a primate bacteremia model. J Infect Dis 164(2):383–8
Reyes AL, Kim Y (2022) Optimal regulation of tumor-associated neutrophils in lung cancer progression. R Soc Open Sci 9:210,705
Roussos E, Condeelis J, Patsialou A (2011) Chemotaxis in cancer. Nat Rev Cancer 11(8):573–587
Ruhland M, Coussens L, Stewart S (2016a) Senescence and cancer: an evolving inflammatory paradox. Biochim Biophys Acta 1865(1):14–22
Ruhland M, Loza A, Capietto A, Luo X, Knolhoff B, Flanagan K, Belt B, Alspach E, Leahy K, Luo J, Schaffer A, Edwards J, Longmore G, Faccio R, DeNardo D, Stewart S (2016b) Stromal senescence establishes an immunosuppressive microenvironment that drives tumorigenesis. Nat Commun 7:11762
Salmon H, Franciszkiewicz K, Damotte D, Dieu-Nosjean M, Validire P, Trautmann A, Mami-Chouaib F, Donnadieu E (2012) Matrix architecture defines the preferential localization and migration of t cells into the stroma of human lung tumors. J Clin Invest 122(3):899–910
Selivanova G, Wiman K (2007) Reactivation of mutant p53: molecular mechanisms and therapeutic potential. Oncogene 26(15):2243–54
Serrano M, Lin A, McCurrach M, Beach D, Lowe S (1997) Oncogenic RAS provokes premature cell senescence associated with accumulation of p53 and p16ink4a. Cell 88(5):593–602
Siegel R, Miller K, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68(1):7–30
Singh A, Arya R, Trivedi A, Sanyal S, Baral R, Dormond O, Briscoe D, Datta D (2013) Chemokine receptor trio: CXCR3, CXCR4 and CXCR7 crosstalk via CXCL11 and CXCL12. Cytokine Growth Factor Rev 24(1):41–9
Song J, Chang C, Wu C, Dinh T, Jan J, Huang K, Chou M, Shiue T, Yeh K, Ke Y, Yeh T, Ta Y, Lee C, Huang J, Sung Y, Shia K, Chen Y (2021) A highly selective and potent CXCR4 antagonist for hepatocellular carcinoma treatment. Proc Natl Acad Sci USA 118(13):e2015433,118
Stokes CL, Lauffenburger DA (1991) Analysis of the roles of microvessel endothelial cell random motility and chemotaxis in angiogenesis. J Theor Biol 152:377–403
Stolarska M, Kim Y, Othmer H (2009) Multiscale models of cell and tissue dynamics. Philos Trans R Soc A 367:3525–3553
Suveges S, Eftimie R, Trucu D (2022) Re-polarisation of macrophages within collective tumour cell migration: a multiscale moving boundary approach. Front Appl Math Stat 7:799,650
Szatmary A, Stuelten C, Nossal R (2014) Improving the design of the agarose spot assay for eukaryotic cell chemotaxis. RSC Adv 4(100):57343–57349
Szpakowska M, Nevins A, Meyrath M, Rhainds D, Dhuys T, Guite-Vinet F, Dupuis N, Gauthier P, Counson M, Kleist A, St-Onge G, Hanson J, Schols D, Volkman B, Heveker N, Chevigne A (2018) Different contributions of chemokine n-terminal features attest to a different ligand binding mode and a bias towards activation of ACKR3/CXCR7 compared with CXCR4 and CXCR3. Br J Pharmacol 175(9):1419–1438
Takekoshi T, Ziarek J, Volkman B, Hwang S (2012) A locked, dimeric CXCL12 variant effectively inhibits pulmonary metastasis of CXCR4-expressing melanoma cells due to enhanced serum stability. Mol Cancer Ther 11(11):2516–25
Taromi S, Kayser G, Catusse J, von Elverfeldt D, Reichardt W, Braun F, Weber W, Zeiser R, Burger M (2016) CXCR4 antagonists suppress small cell lung cancer progression. Oncotarget 7(51):85185–85195
Tasheva S, Ivanova T, Popova V, Iliev I, Stankov S, Fidan H, Mazova N, Stoyanova A (2019) Coefficient of diffusion of tannins in extracts from physalis leaves (Physalis peruviana l.). Bul Chem Commun 51:209–213
Taverna S, Amodeo V, Saieva L, Russo A, Giallombardo M, Leo GD, Alessandro R (2014) Exosomal shuttling of miR-126 in endothelial cells modulates adhesive and migratory abilities of chronic myelogenous leukemia cells. Mol Cancer 13:169
Tchou J, Zhao Y, Levine B, Zhang P, Davis M, Melenhorst J, Kulikovskaya I, Brennan A, Liu X, Lacey S, Williams A, So A, Conejo-Garcia J, Plesa G, Young R, McGettigan S, Campbell J, Pierce R, Matro J, DeMichele A, Clark A, Cooper L, Schuchter L, Vonderheide R, June C (2017) Safety and efficacy of intratumoral injections of chimeric antigen receptor (CAR) T cells in metastatic breast cancer. Cancer Immunol Res 5(12):1152–1161
Topalian S, Hodi F, Brahmer J, Gettinger S, Smith D, McDermott D, Powderly J, Carvajal R, Sosman J, Atkins M, Leming P, Spigel D, Antonia S, Horn L, Drake C, Pardoll D, Chen L, Sharfman W, Anders R, Taube J, McMiller T, Xu H, Korman A, Jure-Kunkel M, Agrawal S, McDonald D, Kollia G, Gupta A, Wigginton J, Sznol M (2012) Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 366(26):2443–54
Toritsuka M, Kimoto S, Muraki K, Landek-Salgado M, Yoshida A, Yamamoto N, Horiuchi Y, Hiyama H, Tajinda K, Keni N, Illingworth E, Iwamoto T, Kishimoto T, Sawa A, Tanigaki K (2013) Deficits in microRNA-mediated Cxcr4/Cxcl12 signaling in neurodevelopmental deficits in a 22q11 deletion syndrome mouse model. Proc Natl Acad Sci USA 110(43):17552–17557
Tyson R, Stern L, LeVeque R (2000) Fractional step methods applied to a chemotaxis model. J Math Biol 41(5):455–75
Uy G, Rettig M, Motabi I, McFarland K, Trinkaus K, Hladnik L, Kulkarni S, Abboud C, Cashen A, Stockerl-Goldstein K, Vij R, Westervelt P, DiPersio J (2012) A phase 1/2 study of chemosensitization with the cxcr4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood 119(17):3917–24
Veldkamp C, Ziarek J, Su J, Basnet H, Lennertz R, Weiner J, Peterson F, Baker J, Volkman B (2009) Monomeric structure of the cardioprotective chemokine SDF-1/CXCL12. Protein Sci 18(7):1359–69
Waldman A, Fritz J, Lenardo M (2020) A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol 20(11):651–668
Welch D, Schissel D, Howrey R, Aeed P (1989) Tumor-elicited polymorphonuclear cells, in contrast to normal circulating polymorphonuclear cells, stimulate invasive and metastatic potentials of rat mammary adenocarcinoma cells. Proc Natl Acad Sci USA 86(15):5859–63
Wong W, Lo A, Siu L, Leung J, Tu S, Tai S, Lam S, Wong K (2013) Reference ranges for lymphocyte subsets among healthy Hong Kong Chinese adults by single-platform flow cytometry. Clin Vaccine Immunol 20(4):602–6
Woodcock E, Land S, Andrews R (1993) A low affinity, low molecular weight endothelin—a receptor present in neonatal rat heart. Clin Exp Pharmacol Physiol 20(5):331–4
Yu T, Liu K, Wu Y, Fan J, Chen J, Li C, Yang Q, Wang Z (2014) MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/\(\beta \)-catenin signaling pathway. Oncogene 33(42):5017–27
Yuan A, Hsiao Y, Chen H, Chen H, Ho C, Chen Y, Liu Y, Hong T, Yu S, Chen J, Yang P (2015) Opposite effects of m1 and m2 macrophage subtypes on lung cancer progression. Sci Rep 5:14,273
Yuan K, Singh R, Rezonzew G, Siegal G (2006) Cell motility in cancer invasion and metastasis, cancer metastasis—biology and treatment, in vitro matrices for studying tumor cell invasion, vol 8, ECM, Springer, Cham, pp 25–54
Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, Li C, Chong M, Ibrahim T, Mercatali L, Amadori D, Lu X, Xie D, Li Q, Wang X (2013) miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol 15(3):284–94
Zhou W, Guo S, Liu M, Burow M, Wang G (2019) Targeting CXCL12/CXCR4 axis in tumor immunotherapy. Curr Med Chem 26(17):3026–3041
Ziarek J, Veldkamp C, Zhang F, Murray N, Kartz G, Liang X, Su J, Baker J, Linhardt R, Volkman B (2013) Heparin oligosaccharides inhibit chemokine (CXC motif) ligand 12 (CXCL12) cardioprotection by binding orthogonal to the dimerization interface, promoting oligomerization, and competing with the chemokine (CXC motif) receptor 4 (CXCR4) N terminus. J Biol Chem 288(1):737–46
Zou W (2022) Immune regulation in the tumor microenvironment and its relevance in cancer therapy. Cell Mol Immunol 19(1):1–2
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2021R1A2C1010891) (Y.K.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2021R1A2C1010891) (Y.K.).
Appendices
Appendix
Parameter estimation
1.1 Parameter values
\(D_T\) (Diffusion coefficient of T cell): From the measured value of the random motility of T cells in a nest of the tumor in Salmon et al. (2012), Li et al. calculated the motility coefficient of 5 \(\upmu \)m\(^2/\)min (Li et al. 2019). We take \(D_T = 5\,\upmu \)m\(^2/\)min \(= 8.33\times 10^{-10}\) cm\(^2/\)s.
\(D_n, D_{ns}\) (Diffusion coefficient of tumor cells and STCs): Measured random motility of typical tumor cells was used in various experimental and theoretical studies including (Kim et al. 2018b, 2019b). We take this value and assume that both tumor cells and STCs have the same diffusion coefficient, leading to \(D_n=D_{ns} = 1.0\times 10^{-11}\) cm\(^2/\)s.
\(D_L\) (Diffusion coefficient of CXCL12): Veldkamp et al. (2009) obtained the measurement of the self-diffusion coefficient of CXCL12 in an experiment involving a water-suppressed longitudinal coding-decoding (water-sLED) system. The diffusion constant of CXCL12 in water was measured to be \(1.74\times 10^{-6}\) cm\(^2/\)s in Szatmary et al. (2014) and Veldkamp et al. (2009). We take \(D_L = 1.74\times 10^{-6}\) cm\(^2/\)s.
\(D_A\) (Diffusion coefficient of CXCL12 antibody): In an experimental study of the inhibitory effect of tannic acid on CXCL12/CXCR4 axis (Chen et al. 2003), the diffusion coefficient of tannin, the CXCL12 antibody, has been reported to be in a range of (3.8–18.4) \(\times 10^{-5}\) cm\(^2/\)s (Mhessn et al. 2011). On the other hand, the diffusion coefficient of tannin was measured to be [6.89 \(\times 10^{-11}\)–1.66 \(\times 10^{-4}\)) cm\(^2/\)s in various studies Tasheva et al. (2019)] [also see refs in Tasheva et al. (2019)]. We take \(D_A = 3.8\times 10^{-7}\) cm\(^2/\)s.
\(\mu _{L}\) (Decay rate of CXCL12): The half-life of CXCL12 was reported to be around 26.5 ± 8.7 min (Takekoshi et al. 2012; Ziarek et al. 2013). In addition, the half-life of various types of CXCL were reported to be 2–15.5 h (Orlikowsky et al. 2004; Redl et al. 1991b, a). By taking 26.5 min for the half-life, we get \(\mu _L = ln(2)/(26.5/60)\) h\(^{-1} = 1.5694\times 10^{-1}\) h\(^{-1} = 4.36 \times 10^{-4}\) s\(^{-1}\).
\(\mu _{D}\) (Decay rate of binding inhibitors): The half-life of AMD3100, inhibitor of CXCL12-CXCR4 binding, was reported to be 3.6\(\;h\) (Hendrix et al. 2000). On the other hand, the half-life was measured to be 1\(\;h\) in mice after injecting AMD3100 (6 mg/kg, SC). By taking the half-life of 3.6 h, we get \(\mu _{D} = ln(2)/(3.6)\) h\(^{-1} = 5.35 \times 10^{-5}\) s\(^{-1}\).
\(\mu _{A}\) (Decay rate of CXCL12 inhibitors): The half-life of CXCL12 inhibitors such as tannic acid was estimated to be 0.96 h (Chen et al. 2003), leading to the decay rate \(\mu _{A} = ln(2)/(0.96)\) h\(^{-1} = 2.01\times 10^{-4}\) s\(^{-1}\).
\(k_{-1}\) (Dissociation rate): Larson et al. (2015) calculated the dissociation rate of CXCL12 to be about (0.031–0.045) s\(^{-1}\) by adding more than 100 times AMD3100 to the previous mixture. We take \(k_{-1} = 0.031\) s\(^{-1}\).
\(k_{1}\) (Association rate): Larson et al. (2015) calculated the \(K_D\) value of the labeled ligand from the HTRF ratio generated after reaching binding equilibrium in response to different concentrations of ligands. In the saturated binding empirical system, the binding activities increase as the concentration of labeled CXCL12 increases. The level of ligands binding to half of the receptor site at equilibrium or \(K_D\) was 29.3 nM. We take \(K_D = 29.3\) nM, leading to the association rate \(k_1 = \frac{k_{-1}}{K_D} = 0.011\) nM\(^{-1}\)s\(^{-1}\).
\(r, r_s\) (Proliferation rates of tumor cells): We take the proliferation rate of non-senescent tumor cells, \(r = 3.33\times 10^{-5}\) s\(^{-1}\), based on experimental observations in (Choi et al. 2021). We assume that senescent tumor cells have a lower growth rate than non-senescent cells. Thus, we take \(r_s = 2.22\times 10^{-5}\) s\(^{-1}\).
\(\lambda _L,\lambda _s\) (Production rate of CXCL12 from two types of tumor cells): Choi et al. (2021) observed that the cytokine shield was formed when the value of CXCL12 was high (> 20 times), and STCs secrete more CXCL12 than non-senescent tumor cells (\(\lambda _L < \lambda _s\)). Thus, we set the CXCL12 production rates, \(\lambda _L=2.78 \times 10^{-1}\) nM cm\(^3\) g\(^{-1}\) s\(^{-1}\) for non-senescent tumor cells and \(\lambda _s = 30 \lambda _L = 8.33\) nM cm\(^3\) g\(^{-1}\) s\(^{-1}\) for STCs.
\(K_\delta , K_2\) (Hill-type coefficients in CXCL12-induced switching): Choi et al. (2021) showed that the overexpressed CXCL12 (> 20-fold) can downregulate the CXCR4 activities and inhibit the chemotaxis of T cells, forming the cytokine shield. In this work, this suppression is controlled by the inhibition term \(\delta _L(L) = \bigg ( \delta _{0} + \delta _{1} \frac{L^m}{K_\delta ^m+ L^m} \bigg )\) in the chemotaxis term of the form \(\frac{\partial T}{\partial t} = -\nabla \cdot \bigg ( \chi _L([\overline{L \cdot R}]) T \frac{\nabla L}{\delta _L+ \sigma _L |\nabla L| } \bigg )\) in Eq. (6) and the degradation of CXCR4 in response to the high CXCL12 level is regulated in the L-dependent degradation term \(k_{2}(L) = k_{20} \frac{L^{n_2}}{K_2^{n_2}+ L^{n_2}}\) in Eq. (11). Based on these experimental results, we assume that the suppression of the chemotactic movement and degradation of CXCR4 of T cells are activated when the CXCL12 level exceeds 7 times the reference value. Thus, we take \(K_\delta = K_2 = 700\) nM.
1.2 Reference concentrations
T cells (\(T^*\)): Wong et al. reported the 95\(\%\) range for the absolute counts of CD8+ T cells in several regional groups: (224–1014) cells/\(\upmu \)l (Hong Kong), (201–931) cells/\(\upmu \)l (Beijing), (243–1206) cells/\(\upmu \)l (Asian), (170–880) cells/\(\upmu \)l (European), and (145–884) cells/\(\upmu \)l (African) (Wong et al. 2013). We take \(T^* = 1000\) cells/\(\upmu \)l \(= 10^6\) cells/cm\(^3\).
Tumor cells (\(n^*, n_s^*\)): We assume that tumor cells are uniformly distributed in the tumor. We also take same reference values for two types of tumor cells (tumor cells and STCs), leading to \(n^* = n_s^* = 10^9~\)cells/cm\(^3\) as in ODonoghue et al. (1995), Friedman et al. (2006) and Kim et al. (2018b, 2019b).
CXCL12 (\(L^*\)) and CXCL12 antibody (\(A^*\)): Larson et al. (2015) performed an associative binding experiment using a range of the CXCL12 concentration, 0–100 nM. The steady state of the complex variables in the associated experiment was positively correlated with the CXCL12 input. Chen et al. (2003) reported that the inhibitory effect of CXCL12 antibody (tannic acid) on CXCL12-induced cell migration depends on dose, with an \(IC_{50} = 7.5~\upmu \)g/ml. In this experiment of the CXCL12/CXCR4 axis, tannic acid (10 \(\upmu \)g/ml) was shown to block cell migration in the presence of CXCL12 (100 ng/ml) in a chemotaxis chamber assay. Aqueous and organic extracts of Lianquiao were able to inhibit the CXCL12-induced cell migration (Chen et al. 2003). For example, organic extracts of Lianquiao can fully inhibit the chemotactic movement of CXCL12-induced infiltration at a concentration 200 \(\upmu \)g/ml. In a study of CXCL12-induced T cell infiltration (Choi et al. 2021), CXCL12 neutralizing antibody of the concentration of 5 \(\upmu \)g/ml was applied to the medium in the lower chamber of boyden chamber system for 24 h. Median serum levels of CXCL12 in control and various cancer patients were 0.865 ng/ml (control), 1.277 ng/ml (esophageal cancer), 1.118 ng/ml (adenocarcinoma of esophagus), and 1.501 ng/ml (squamous cell cancer of esophagus) (Lukaszewicz-Zajac and Mroczko 2016). In a study of CXCL12-CXCR4 axis, Song et al. (2021) reported that CXCL12 of concentration 10 nM can enhance the invasiveness of HCA-1 cells (> fivefold) relative control (CXCL12-) but its inhibitor BPRCX807 can negate this aggressive migration by 25% and 80% with inhibitor concentrations of 100, 1000 nM, respectively. We take \(A^* = 7.5 \times 10^{-6}\) g/cm\(^3\) and \(L^* = 100\) nM.
CXCR4 (\([R]^*\)), CXCL12-CXCR4 complex (\([\overline{L \cdot R}]^*\)): Median serum levels of CXCR4 in control and various cancer patients were 0.932 ng/ml (control), 0.443 ng/ml (esophageal cancer), 0.403 ng/ml (adenocarcinoma of esophagus), and 0.534 ng/ml (squamous cell cancer of esophagus) (Lukaszewicz-Zajac and Mroczko 2016), while the \(IC_{50}\) of CXCR4 antagonist has been reported to be in a range of (2.1–23.8) nM (Hanes et al. 2015; Hoffmann et al. 2012; Szpakowska et al. 2018). Considering the average values of CXCL12 concentration relative to the median CXCR4 concentration in the control group (Lukaszewicz-Zajac and Mroczko 2016), we take \([R]^* = 100\) nM. We take the same reference value for the CXCL12-CXCR4 complex, \([\overline{L \cdot R}]^* = 100\) nM.
The computational algorithm in the multi-scale model
-
Step 0. Initialization.
-
Step 0.1. Fix a uniform grid for \(\varOmega \) and initialize concentrations of CXCL12 (L) and densities of non-senescent tumor cells (n) and STCs (\(n_s\)) at each lattice point.
-
Step 0.2. Set the locations of the blood vessels outside the tumor based on the spatial distribution of tumor cell density (\(n,n_s\)). Initialize cell-based component by randomly placing T cells outside the tumor.
-
Step 0.3. Set the initial condition of the receptor (\([R]_i(t)\)) and receptor complex \([\overline{L \cdot R}]_i(t)\) at each T cell i.
-
-
Step 1. Generate discrete T cells at the blood vessels in a random time and random location of blood vessels and set initial conditions for the receptor and receptor complex at the T cell i.
-
Step 2. Update the CXCL12 concentration at each T cell by interpolation from grid point to the cell site (see Dallon and Othmer 1997; Kim et al. 2011, 2018a; Kim and Othmer 2015). Solve the intracellular receptor binding equations (13)–(14) to update the receptor complex value at each T cell i.
-
Step 3. Use the level of the receptor complex (\([\overline{L \cdot R}]_i(t)\)) in order to determine movement of T cell i based on the switching behavior \(\chi _0 \bigg ( \frac{[\overline{L \cdot R}]^{n_1}}{K_\chi ^{n_1}+ [\overline{L \cdot R}]^{n_1}} \bigg )\) in Eq. (4) and gradient of CXCL12, \(\bigg ( \dfrac{ \nabla L}{\delta _L(L)+\sigma _L |\nabla L| } \bigg )\).
-
Step 4. Determine the direction of motion for a T cell when its invasion switch is activated. The migration direction is based on up-gradient of CXCL12 (\(\nabla L\)) in the chemotaxis mode and randomly determined with a random number in the random walk mode.
-
Step 5. Translation of T cells: Update the location of T cells based on mobility switch and movement direction in Steps 3–4.
-
Step 6. Solve the reaction–diffusion equations (7), (8) and (15) of densities of non-senescent tumor cells and STCs, and CXCL concentration on a regular grid, using the ADI method, lagging the consumption term. Interpolation of receptor and receptor complex from the cell site to grid point is done as in Dallon and Othmer (1997), Kim et al. (2011, 2018a) and Kim and Othmer (2015). Replace Eq. (15) with Eqs. (23)–(24) in the presence of CXCL12 antibody for therapeutic approaches.
-
Step 7. Go to Step 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, Y., Lee, J., Lee, C. et al. Role of senescent tumor cells in building a cytokine shield in the tumor microenvironment: mathematical modeling. J. Math. Biol. 86, 14 (2023). https://doi.org/10.1007/s00285-022-01850-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00285-022-01850-z