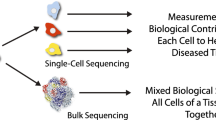
Abstract
Childhood cancer is the second leading cause of death in children aged 1 to 14. Although survival rates have vastly improved over the past 40 years, cancer resistance and relapse remain a significant challenge. Advances in single-cell technologies enable dissection of tumors to unprecedented resolution. This facilitates unraveling the heterogeneity of childhood cancers to identify cell subtypes that are prone to treatment resistance. The rapid accumulation of single-cell data from different modalities necessitates the development of novel computational approaches for processing, visualizing, and analyzing single-cell data. Here, we review single-cell approaches utilized or under development in the context of childhood cancers. We review computational methods for analyzing single-cell data and discuss best practices for their application. Finally, we review the impact of several studies of childhood tumors analyzed with these approaches and future directions to implement single-cell studies into translational cancer research in pediatric oncology.


Similar content being viewed by others
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics, 2021. CA Cancer J Clin 71:7–33
Neaga A et al (2021) Why do children with acute lymphoblastic leukemia fare better than adults?. Cancers (Basel) 13
Lee SHR, Li Z, Tai ST, Oh BLZ, Yeoh AEJ (2021) Genetic alterations in childhood acute lymphoblastic leukemia: interactions with clinical features and treatment response. Cancers (Basel) 13
Lee SHR, Li Z, Tai ST, Oh BLZ, Yeoh AEJ (2021) Genetic alterations in childhood acute lymphoblastic leukemia: interactions with clinical features and treatment response. Cancers (Basel) 13:4068
Radtke I et al (2009) Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci U S A 106:12944–12949
Aynaud MM et al (2020) Transcriptional programs define intratumoral heterogeneity of Ewing sarcoma at single-cell resolution. Cell Rep 30:1767-1779.e1766
Bandura DR et al (2009) Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem 81:6813–6822
Ornatsky O et al (2010) Highly multiparametric analysis by mass cytometry. J Immunol Methods 361:1–20
Leelatian N et al (2017) Single cell analysis of human tissues and solid tumors with mass cytometry. Cytometry B Clin Cytom 92:68–78
Jaimes MC et al (2021) Full spectrum flow cytometry and mass cytometry: a 32-marker panel comparison. Cytometry A
Jager A, Sarno J, Davis KL (2021) Mass cytometry of hematopoietic cells. Methods Mol Biol 2185:65–76
Spitzer MH, Nolan GP (2016) Mass cytometry: single cells, many features. Cell 165:780–791
Good Z et al (2018) Single-cell developmental classification of B cell precursor acute lymphoblastic leukemia at diagnosis reveals predictors of relapse. Nat Med 24:474–483
Angelo M et al (2014) Multiplexed ion beam imaging of human breast tumors. Nat Med 20:436–442
Giesen C et al (2014) Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 11:417–422
Black S et al (2021) CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc 16:3802–3835
Goltsev Y et al (2018) Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174:968-981.e915
Keren L et al (2018) A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174:1373-1387.e1319
Keren L et al (2019) MIBI-TOF: a multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci Adv 5:eaax5851
Liu, C.C. et al. Reproducible, high-dimensional imaging in archival human tissue by multiplexed ion beam imaging by time-of-flight (MIBI-TOF). Laboratory Investigation (2022).
Liu CC et al (2022) Multiplexed ion beam imaging: insights into pathobiology. Annu Rev Pathol 17:403–423
Kammersgaard MB et al (2020) Abstract PO-041: Multiplexed ion beam imaging to describe tumor-immune microenvironment and tumor heterogeneity in neuroblastoma. Cancer Res 80:PO-041-PO-041
Batth IS et al (2020) Rare osteosarcoma cell subpopulation protein array and profiling using imaging mass cytometry and bioinformatics analysis. BMC Cancer 20:715
Gerdtsson E et al (2018) Multiplex protein detection on circulating tumor cells from liquid biopsies using imaging mass cytometry. Converg Sci Phys Oncol 4
Black S et al (2021) CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc 16:3802–3835
Bosisio FM et al (2022) Next-generation pathology using multiplexed immunohistochemistry: mapping tissue architecture at single-cell level. Front Oncol 12:918900
Cesano A, Marincola FM, Thurin M (2020) Status of immune oncology: challenges and opportunities. Methods Mol Biol 2055:3–21
Gawad C, Koh W, Quake SR (2014) Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci 111:17947–17952
Mehtonen J et al (2020) Single cell characterization of B-lymphoid differentiation and leukemic cell states during chemotherapy in ETV6-RUNX1-positive pediatric leukemia identifies drug-targetable transcription factor activities. Genome Med 12:99
Caron M et al (2020) Single-cell analysis of childhood leukemia reveals a link between developmental states and ribosomal protein expression as a source of intra-individual heterogeneity. Sci Rep 10:8079
Louka E et al (2021) Heterogeneous disease-propagating stem cells in juvenile myelomonocytic leukemia. J Exp Med 218
Hovestadt V et al (2019) Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 572:74–79
Vladoiu MC et al (2019) Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572:67–73
Gillen AE et al (2020) Single-cell RNA sequencing of childhood ependymoma reveals neoplastic cell subpopulations that impact molecular classification and etiology. Cell Rep 32:108023
Hovestadt V et al (2019) Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 572:74–79
Jansky S et al (2021) Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nat Genet 53:683–693
Corces MR et al (2017) An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14:959–962
Shahi P, Kim SC, Haliburton JR, Gartner ZJ, Abate AR (2017) Abseq: ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Sci Rep 7:44447
Stoeckius M et al (2017) Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14:865–868
Mimitou EP et al (2019) Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat Methods 16:409–412
Hao Y et al (2021) Integrated analysis of multimodal single-cell data. Cell 184:3573-3587.e3529
Bai Z et al (2022) Single-cell antigen-specific landscape of CAR T infusion product identifies determinants of CD19-positive relapse in patients with ALL. Sci Adv 8:2820
Lee J, Hyeon DY, Hwang D (2020) Single-cell multiomics: technologies and data analysis methods. Exp Mol Med 52:1428–1442
Mizuno H, Tsuyama N, Date S, Harada T, Masujima T (2008) Live single-cell metabolomics of tryptophan and histidine metabolites in a rat basophil leukemia cell. Anal Sci 24:1525–1527
Pan N, Rao W, Yang Z (2020) Single-probe mass spectrometry analysis of metabolites in single cells. Methods Mol Biol 2064:61–71
Ahl PJ et al (2020) Met-Flow, a strategy for single-cell metabolic analysis highlights dynamic changes in immune subpopulations. Commun Biol 3:305
Arguello RJ et al (2020) SCENITH: a flow cytometry-based method to functionally profile energy metabolism with single-cell resolution. Cell Metab 1063–1075:e1067
Sharick JT et al (2020) Metabolic heterogeneity in patient tumor-derived organoids by primary site and drug treatment. Front Oncol 10:553
McCarthy DJ, Campbell KR, Lun AT, Wills QF (2017) Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33:1179–1186
Satija R, Farrell JA, Gennert D, Schier AF, Regev A (2015) Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33:495–502
Wolf FA, Angerer P, Theis FJ (2018) SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19:15
Palla G et al (2022) Squidpy: a scalable framework for spatial omics analysis. Nat Methods 19:171–178
Gayoso A et al (2022) A Python library for probabilistic analysis of single-cell omics data. Nat Biotechnol 40:163–166
Nowicka M et al (2017) CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. Res 6:748
Chen H et al (2016) Cytofkit: a bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput Biol 12:e1005112
Greenwald NF et al (2022) Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat Biotechnol 40:555–565
Luecken MD, Theis FJ (2019) Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol 15:e8746
Kotecha N, Krutzik PO, Irish JM (2021) Web-based analysis and publication of flow cytometry experiments. Current protocols in cytometry Chapter 10, Unit10.17-Unit10.17
Amir E-AD et al (2019) Development of a comprehensive antibody staining database using a standardized analytics pipeline. Front Immunol 10:1315–1315
Belkina AC et al (2019) Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat Commun 10:5415
Lo YC et al (2022) CytofIn enables integrated analysis of public mass cytometry datasets using generalized anchors. Nat Commun 13:934
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420
Baek S, Lee I (2020) Single-cell ATAC sequencing analysis: from data preprocessing to hypothesis generation. Comput Struct Biotechnol J 18:1429–1439
Kopp W, Akalin A, Ohler U (2022) Simultaneous dimensionality reduction and integration for single-cell ATAC-seq data using deep learning. Nature Machine Intelligence 4:162–168
Tran HTN et al (2020) A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol 21:12
Hardoon DR, Shawe-Taylor J (2011) Sparse canonical correlation analysis. Mach Learn 83:331–353
Argelaguet R et al (2018) Multi-omics factor analysis-a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol 14:e8124
Welch JD et al (2019) Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177:1873-1887.e1817
Witkowski MT et al (2020) Extensive remodeling of the immune microenvironment in b cell acute lymphoblastic leukemia. Cancer Cell 37:867-882.e812
Avila Cobos F, Alquicira-Hernandez J, Powell JE, Mestdagh P, De Preter K (2020) Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat Commun 11:5650
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA (2018) Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 1711:243–259
Campana D (2010) Minimal residual disease in acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program 2010:7–12
van der Velden VH, Boeckx N, van Wering ER, van Dongen JJ (2004) Detection of minimal residual disease in acute leukemia. J Biol Regul Homeost Agents 18:146–154
Zhang Y et al (2022) Elucidating minimal residual disease of paediatric B-cell acute lymphoblastic leukaemia by single-cell analysis. Nat Cell Biol 24:242–252
Acknowledgements
We thank all members of the Davis lab for helpful discussions.
Funding
KLD is the Anne T. and Robert M. Bass Endowed Faculty Scholar in Pediatric Cancer and Blood Diseases. This work is supported by Stanford Maternal and Child Health Research Institute, NCI U54 CA232568, NCI R01 CA251858, NCI R21 CA234529, NCI R01 CA251858-01A1S1, Mark Foundation Aspire Award, The Andrew McDonough B Positive Foundation, W81XWH-19-PRCRP-CDA Department of Defense Young Investigator Award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is a contribution to the special issue on: Single-cell and spatial multi-omics in clinical outcomes studies - Guest Editor: Brice Gaudillière
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lo, YC., Liu, Y., Kammersgaard, M. et al. Single-cell technologies uncover intra-tumor heterogeneity in childhood cancers. Semin Immunopathol 45, 61–69 (2023). https://doi.org/10.1007/s00281-022-00981-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-022-00981-1