Abstract
Purpose
Glioma is the most common primary tumor in the brain, accounting for 81% of intracranial malignancies. Nowadays, cancer immunotherapy has become a novel and revolutionary treatment for patients with advanced, highly aggressive tumors. However, to date, there are no effective biomarkers to reflect the response of glioma patients to immunotherapy. In this study, we aimed to assess the clinical predictive value of ITGB2 in patients with glioma.
Methods
The correlation between ITGB2 expression levels and glioma progression was explored and validated using data from CGGA, TCGA, GEO datasets, and patient samples from our hospital. Univariate and multivariate cox regression models were developed to determine the predictive role of ITGB2 on the prognosis of patients with glioma. The relationship between ITGB2 and immune activation was then analyzed. Finally, we predicted the immunotherapy response in both high and low ITGB2 expression subgroups.
Results
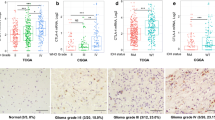
ITGB2 was significantly elevated in gliomas with higher malignancy and predicted poor prognosis. In multivariate analysis, the hazard ratio for ITGB2 expression (low versus high) was 0.71 with 95% CI (0.59–0.85) (P < 0.001). Furthermore, we found that ITGB2 stratified glioma patients into high and low ITGB2 expression subgroups, exhibiting different clinical outcomes and immune activation status. At last, we demonstrated that glioma patients with high ITGB2 expression levels had better immunotherapy response.
Conclusions
This study demonstrated ITGB2 as a novel predictor for clinical prognosis and response to immunotherapy in gliomas. Assessing expression levels of ITGB2 is a promising method to discover patients that may benefit from immunotherapy.






Similar content being viewed by others
Data availability
Publicly available datasets were analyzed in this study. These data can be found here: http://gliovis.bioinfo.cnio.es/ and http://tisch.comp-genomics.org/home/. The supplementary material for this article can be found online. All processed data and R codes used in this study can be obtained from the corresponding author on reasonable request.
Abbreviations
- ACC:
-
Adrenocortical carcinoma
- BLCA:
-
Bladder urothelial carcinoma
- BP:
-
Biological process
- BRCA:
-
Breast invasive carcinoma
- CC:
-
Cellular component
- CESC:
-
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CGGA:
-
Chinese glioma genome atlas
- CHOL:
-
Cholangiocarcinoma
- COAD:
-
Colon adenocarcinoma
- CTLA4:
-
Cytotoxic T-lymphocyte-associated protein 4
- DEG:
-
Differential expressed gene
- DLBC:
-
Lymphoid neoplasm diffuse large B-cell lymphoma
- DSS:
-
Disease-specific survival
- ESCA:
-
Esophageal carcinoma
- GBM:
-
Glioblastoma multiforme
- GEO:
-
Gene expression omnibus
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- HNSC:
-
Head and neck squamous cell carcinoma
- ImmuCellAI:
-
Immune cells abundance identifier
- ICB:
-
Immune checkpoint blockades
- IDO1:
-
Indoleamine 2,3-dioxygenase
- IHC:
-
Immunohistochemistry
- ITGB2:
-
Integrin beta-2
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- KICH:
-
Kidney chromophobe
- KIRC:
-
Kidney renal clear cell carcinoma
- KIRP:
-
Kidney renal papillary cell carcinoma
- LCML:
-
Chronic myelogenous leukemia
- LGG:
-
Brain lower-grade glioma
- LIHC:
-
Liver hepatocellular carcinoma
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- MESO:
-
Mesothelioma
- MF:
-
Molecular function
- OS:
-
Overall survival
- OV:
-
Ovarian serous cystadenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PCPG:
-
Pheochromocytoma and paraganglioma
- PD-1:
-
Programmed cell death 1
- PDL-1 (CD274):
-
Programmed cell death 1 ligand 1
- PFI:
-
Progression-free interval
- PRAD:
-
Prostate adenocarcinoma
- READ:
-
Rectum adenocarcinoma
- SARC:
-
Sarcoma
- SKCM:
-
Skin cutaneous melanoma
- SubMap:
-
Subclass mapping
- STAD:
-
Stomach adenocarcinoma
- TCGA:
-
The cancer genome atlas
- TGCT:
-
Testicular germ cell tumors
- THCA:
-
Thyroid carcinoma
- THYM:
-
Thymoma
- TIICs:
-
Tumor-infiltrating immune cells
- TIGIT:
-
T cell immunoreceptor with Ig and ITIM domains
- TIM3 (HAVCR2):
-
(T-cell immunoglobulin and mucin domain-containing protein 3/hepatitis A virus cellular receptor 2)
- TIS:
-
T-cell inflammation
- UCEC:
-
Uterine corpus endometrial carcinoma
- UCS:
-
Uterine carcinosarcoma
- UVM:
-
Uveal melanoma
References
Ostrom QT et al (2018) Epidemiology of intracranial gliomas. Prog Neurol Surg 30:1–11
Ostrom QT et al (2014) The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol 16(7):896–913
Louis DN et al (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114(2):97–109
Bush NAO, Chang SM, Berger MS (2017) Current and future strategies for treatment of glioma. Neurosurg Rev 40(1):1–14
Zeng T, Cui D, Gao L (2015) Glioma: an overview of current classifications, characteristics, molecular biology and target therapies. Front Biosci (Landmark edition) 20:1104–1115
Cui H et al (2016) NF-YC in glioma cell proliferation and tumor growth and its role as an independent predictor of patient survival. Neurosci Lett 631:40–49
Latchman Y et al (2001) PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol 2(3):261–268
Parry RV et al (2005) CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol 25(21):9543–9553
Adorno-Cruz V, Liu H (2019) Regulation and functions of integrin α2 in cell adhesion and disease. Gen Dis 6(1):16–24
Jung S, Yuki K (2016) Differential effects of volatile anesthetics on leukocyte integrin macrophage-1 antigen. J Immunotoxicol 13(2):148–156
Varga G et al (2007) Active MAC-1 (CD11b/CD18) on DCs inhibits full T-cell activation. Blood 109(2):661–669
Lukácsi S et al (2017) The role of CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in complement-mediated phagocytosis and podosome formation by human phagocytes. Immunol Lett 189:64–72
Yakubenko VP, Yadav SP, Ugarova TP (2006) Integrin alphaDbeta2, an adhesion receptor up-regulated on macrophage foam cells, exhibits multiligand-binding properties. Blood 107(4):1643–1650
Bednarczyk M et al (2020) β2 integrins-multi-functional leukocyte receptors in health and disease. Int J Mol Sci 21(4):1402
Cooper J, Giancotti FG (2019) Integrin signaling in cancer: mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 35(3):347–367
Liu M et al (2018) LncRNA ITGB2-AS1 could promote the migration and invasion of breast cancer cells through up-regulating ITGB2. Int J Mol Sci 19(7):1866
Chang CM et al (2013) Innate immunity gene polymorphisms and the risk of colorectal neoplasia. Carcinogenesis 34(11):2512–2520
Ma J et al (2013) Innate immune cell-derived microparticles facilitate hepatocarcinoma metastasis by transferring integrin α(M)β2 to tumor cells. J Immunol (Baltimore, Md.: 1950) 191(6):3453–3461
Almeida J et al (2019) Adipocyte proteome and secretome influence inflammatory and hormone pathways in glioma. Metab Brain Dis 34(1):141–152
Rajaraman P et al (2009) Common variation in genes related to innate immunity and risk of adult glioma. Cancer Epidemiol Biomarkers Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol 18(5):1651–1658
Gladitz J, Klink B, Seifert M (2018) Network-based analysis of oligodendrogliomas predicts novel cancer gene candidates within the region of the 1p/19q co-deletion. Acta Neuropathol Commun 6(1):49
Zhou P et al (2015) No association of VAMP8 gene polymorphisms with glioma in a Chinese han population. Int J Clin Exp Pathol 8(5):5681–5687
Chen Y et al (2015) VAMP8 facilitates cellular proliferation and temozolomide resistance in human glioma cells. Neuro Oncol 17(3):407–418
Wang S, Liu X (2019) The UCSCXENATOOLS R package: a toolkit for accessing genomics data from UCSC xena platform, from cancer multi-omics to single-cell RNA-seq. J Open Sour Softw 4(40):1627
Bowman RL et al (2017) GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol 19(1):139–141
Sun D et al (2020) TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res 49(D1):D1420–D1430
Neftel C et al (2019) An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178(4):835–849
Hänzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14:7
Jia Q et al (2018) Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun 9(1):5361
García-Mulero S et al (2020) Lung metastases share common immune features regardless of primary tumor origin. J Immunother Cancer 8(1):e000491
Li C et al (2021) GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res 49(W1):W242–W246
Yu G et al (2012) Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287
Fang H et al (2019) A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat Genet 51(7):1082–1091
Xu L et al (2018) TIP: a web server for resolving tumor immunophenotype profiling. Can Res 78(23):6575–6580
Auslander N et al (2018) Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med 24(10):1545–1549
Miao YR et al (2020) ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinheim, Baden-Wurttemberg, Germany) 7(7):1902880
Hoshida Y et al (2007) Subclass mapping: identifying common subtypes in independent disease data sets. PloS One 2(11):e1195
Grabowski MM et al (2021) Immune suppression in gliomas. J Neurooncol 151(1):3–12
Varga G et al (2010) LFA-1 contributes to signal I of T-cell activation and to the production of T(h)1 cytokines. J Invest Dermatol 130(4):1005–1012
Zhang X et al (2020) ITGB2-mediated metabolic switch in CAFs promotes OSCC proliferation by oxidation of NADH in mitochondrial oxidative phosphorylation system. Theranostics 10(26):12044–12059
Lanitis E et al (2017) Mechanisms regulating T-cell infiltration and activity in solid tumors. Ann Oncol Off J Eur Soc Med Oncol 28(Suppl_12):xii18–xii32
Baumert BG et al (2016) Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033–26033): a randomised, open-label, phase 3 intergroup study. Lancet Oncol 17(11):1521–1532
Douw L et al (2009) Cognitive and radiological effects of radiotherapy in patients with low-grade glioma: long-term follow-up. Lancet Neurol 8(9):810–818
Dang L, Jin S, Su SM (2010) IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med 16(9):387–397
Laurence MG et al (2017) Oncogenic activities of IDH1/2 mutations: from epigenetics to cellular signaling. Trends Cell Biol 27(10):738–752
Bady P et al (2012) MGMT methylation analysis of glioblastoma on the Infinium methylation beadchip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol 124(4):547–560
van den Bent MJ et al (2013) MGMT-STP27 methylation status as predictive marker for response to PCV in anaplastic oligodendrogliomas and oligoastrocytomas. A report from EORTC study 26951. Clin Cancer Res Off J Am Assoc Cancer Res 19(19):5513–5522
Wick W et al (2013) Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology 81(17):1515–1522
Staedtke V, aDzaye OD, Holdhoff M (2016) Actionable molecular biomarkers in primary brain tumors. Trends Cancer 2(7):338–349
Khan IN et al (2018) Current and emerging biomarkers in tumors of the central nervous system: possible diagnostic, prognostic and therapeutic applications. Semin Cancer Biol 52(1):85–102
Xiao Y et al (2019) Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin Cancer Res Off J Am Assoc Cancer Res 25(16):5002–5014
Chen Y-P et al (2017) Genomic analysis of tumor microenvironment immune types across 14 solid cancer types: immunotherapeutic implications. Theranostics 7(14):3585–3594
Mellman I, Coukos G, Dranoff G (2011) Cancer immunotherapy comes of age. Nature 480(7378):480–489
Gil Del Alcazar CR, Alečković M, Polyak K (2020) Immune escape during breast tumor progression. Cancer Immunol Res 8(4):422–427
Del Paggio JC (2018) Immunotherapy: cancer immunotherapy and the value of cure. Nat Rev Clin Oncol 15(5):268–270
Patel JM et al (2019) Peritumoral administration of DRibbles-pulsed antigen-presenting cells enhances the antitumor efficacy of anti-GITR and anti-PD-1 antibodies via an antigen presenting independent mechanism. J Immunother Cancer 7(1):311
López-Soto A, Gonzalez S, Folgueras AR (2017) IFN signaling and ICB resistance: time is on tumor’s side. Trends Cancer 3(3):161–163
Jiang P et al (2018) Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 24(10):1550–1558
Rotte A (2019) Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J Exp Clin Cancer Res CR 38(1):255
Wei SC et al (2017) Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 170(6):1120-1133.e17
Acknowledgements
The authors would like to thank Zhenyu Song from Fudan University for his generous and outstanding help in this research design. We thank openbiox community and Hiplot team (https://hiplot.com.cn) for providing technical assistance and valuable tools for data analysis and visualization.
Funding
This research was funded by Natural Science Foundation of Shanghai (18ZR1430400).
Author information
Authors and Affiliations
Contributions
HX, AZ and XH collected clinical data. HX, WW performed data analysis. HX, AZ and ML designed and wrote the manuscript. WW and ML contributed to discussion.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethical approval
The study was approved by the Human Investigation Ethical Committee of Shanghai General Hospital, and the written informed consent was obtained from all patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure 1.
ITGB2 expression and correlation with immune signatures in pan-cancer. (a) ITGB2 has a significant relationship with tumor microenvironment in various tumors. Supplementary Figure 2. ITGB2 involved in remodeling tumor immune microenvironment.(a) U-MAP plot demonstrating clustering obtained from single cell RNA sequencing (GSE131928). Clusters annotations: CD8Tex, Mono/Macro, AC-like Malignant, MES-like Malignant, OPC-like Malignant, NPC-like Malignant, Oligodendrocyte. (b-c) ITGB2 specifically expressed in Mono/Macro and CD8Tex. (d-e) GEPIA2021 demonstrated that ITGB2's expression was significantly higher in M2 macrophages compared to M0 and M1 macrophages. Supplementary Figure 3. Gene function enrichment analysis between ITGB2 high and low subgroups. (a) KEGG results for differential expression genes between ITGB2 high and low subgroups. The X-axis represents gene counts and the Y-axis represents different enriched pathways (log2 fold change> 1.5). (b) GO (Gene Ontology) results for differential expression genes (Cut-off criteria for DEGs significance was adj. p value< 0.05 and the absolute value of the log2 fold change> 1.5). The X-axis represents gene ratio and the Y-axis represents different enriched pathways (BP: biological progress; CC: cellular component; MF: molecular function). (c) The plot of top three GO pathways for differential expression genes. (d) Rank-based gene set enrichment analysis shows significantly activated immune related pathways in ITGB2 high subgroup compared with ITGB2 low subgroup (LFC, log fold change). Supplementary Figure 4. Validation of correlation between ITGB2 and immune checkpoints in CGGA and TCGA cohorts. (a) The correlation between PDCD1, CD274, CTLA4 and ITGB2 in TCGA dataset. (b) Correlation between mRNA expression of ITGB2 and PDCD1, CTLA4, TIM3 (HAVCR2), IDO1 and TIGIT in tumor tissues obtained from glioma patients (n = 20). (c) The correlation between TIGIT, IDO1, TIM3(HAVCR2) and ITGB2 in CGGA dataset. (d) The correlation between TIGIT, IDO1, TIM3(HAVCR2) and ITGB2 in TCGA dataset. Supplementary Figure 5. Association between ITGB2 expression and immunotherapeutic response. (a) ROC curves for ITGB2 in predicting the immunotherapy response of glioma patients. (PDF 10377 KB)
Supplementary Sheet 1.
Marker genes of 28 immune cells. Supplementary Sheet 2. Marker genes of 25 immune-related pathways. Supplementary Sheet 3. Marker genes of the steps of the cancer immunity cycle. Supplementary Sheet 4. Marker genes of immunotherapy-predicted pathways. Supplementary Sheet 5. Marker genes of TIICs (Based on ImmuCellAI). Supplementary Sheet 6. Marker genes of TIS scores. Supplementary Sheet 7. GSEA result of ITGB2 high vs low subgroup. (XLSX 66 KB)
Rights and permissions
About this article
Cite this article
Xu, H., Zhang, A., Han, X. et al. ITGB2 as a prognostic indicator and a predictive marker for immunotherapy in gliomas. Cancer Immunol Immunother 71, 645–660 (2022). https://doi.org/10.1007/s00262-021-03022-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-021-03022-2