Abstract
Purpose
Early identification of patients at risk of developing colorectal liver metastases can help personalizing treatment and improve oncological outcome. The aim of this study was to investigate in patients with colorectal cancer (CRC) whether a machine learning-based radiomics model can predict the occurrence of metachronous metastases.
Methods
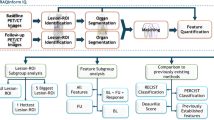
In this multicentre study, the primary staging portal venous phase CT of 91 CRC patients were retrospectively analysed. Two groups were assessed: patients without liver metastases at primary staging, or during follow-up of ≥ 24 months (n = 67) and patients without liver metastases at primary staging but developed metachronous liver metastases < 24 months after primary staging (n = 24). After liver parenchyma segmentation, 1767 radiomics features were extracted for each patient. Three predictive models were constructed based on (1) radiomics features, (2) clinical features and (3) a combination of clinical and radiomics features. Stability of features across hospitals was assessed by the Kruskal–Wallis test and inter-correlated features were removed if their correlation coefficient was higher than 0.9. Bayesian-optimized random forest with wrapper feature selection was used for prediction models.
Results
The three predictive models that included radiomics features, clinical features and a combination of radiomics with clinical features resulted in an AUC in the validation cohort of 86% (95%CI 85–87%), 71% (95%CI 69–72%) and 86% (95% CI 85–87%), respectively.
Conclusion
A machine learning-based radiomics analysis of routine clinical CT imaging at primary staging can provide valuable biomarkers to identify patients at high risk for developing colorectal liver metastases.



Similar content being viewed by others
Abbreviations
- CRC:
-
Colorectal cancer
- CRLM:
-
Colorectal liver metastases
- CEA:
-
Carcinoembryonic antigen
- LM:
-
Liver metastases
- ML:
-
Machine learning
- RF:
-
Random forest
- CI:
-
Confidence interval
References
Haggar FA, Boushey RP (2009) Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg 22:191
Weiss L, Grundmann E, Torhorst J, et al (1986) Haematogenous metastastic patterns in colonic carcinoma: An analysis of 1541 necropsies. J Pathol 150:195–203
Manfredi S, Lepage C, Hatem C, et al (2006) Epidemiology and management of liver metastases from colorectal cancer. Ann Surg 244:254–259
Landreau P, Drouillard A, Launoy G, et al (2015) Incidence and survival in late liver metastases of colorectal cancer. J Gastroenterol Hepatol 30:82–85
Locker GY, Hamilton S, Harris J, et al (2006) ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol 24:5313–5327
Secco GB, Fardelli R, Gianquinto D, et al (1997) Factors influencing incidence and extension of metachronous liver metastases of colorectal adenocarcinoma. A multivariate analysis. Hepatogastroenterology 44:1057–1062
Chuang S-C, Su Y-C, Lu C-Y, et al (2011) Risk factors for the development of metachronous liver metastasis in colorectal cancer patients after curative resection. World J Surg 35:424–429
Rao S-X, Lambregts DM, Schnerr RS, et al (2014) Whole-liver CT texture analysis in colorectal cancer: Does the presence of liver metastases affect the texture of the remaining liver? United European Gastroenterol J 2:530–538
Beckers RCJ, Lambregts DMJ, Schnerr RS, et al (2017) Whole liver CT texture analysis to predict the development of colorectal liver metastases—A multicentre study. Eur J Radiol 92:64–71
Lee SJ, Zea R, Kim DH, et al (2018) CT texture features of liver parenchyma for predicting development of metastatic disease and overall survival in patients with colorectal cancer. Eur Radiol 28:1520–1528
Beckers RCJ, Beets-Tan RGH, Schnerr RS, et al (2017) Whole-volume vs. segmental CT texture analysis of the liver to assess metachronous colorectal liver metastases. Abdom Radiol (NY) 42:2639–2645
Ganeshan B, Miles KA, Young RCD, Chatwin CR (2009) Texture analysis in non-contrast enhanced CT: Impact of malignancy on texture in apparently disease-free areas of the liver. Eur J Radiol 70:101–110
Lambin P, Rios-Velazquez E, Leijenaar R, et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Aerts HJWL, Velazquez ER, Leijenaar RTH, et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Lu W, Chen W (2016) Positron emission tomography/computerized tomography for tumor response assessment-a review of clinical practices and radiomics studies. Transl Cancer Res 5:364–370
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: Images Are More than Pictures, They Are Data. Radiology 278:563–577
Avanzo M, Stancanello J, El Naqa I (2017) Beyond imaging: The promise of radiomics. Phys Med 38:122–139
Beckers R, Lambregts D, Schnerr R, et al (2017) Whole liver CT texture analysis to predict the development of colorectal liver metastases − a multicentre study. European Journal of Cancer 72:S49
Oncoline. https://www.oncoline.nl/. Accessed 25 Jan 2019
Philips - Nederland. In: Philips. https://www.philips.nl/. Accessed 15 Oct 2018
van Griethuysen J. J. M. Fedorov A. Parmar C. Hosny A. Aucoin N. Narayan V. Beets-Tan R. G. H. Fillon-Robin J. C. Pieper S. Aerts HJWL (2017) Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 77(21), e104–e107
Welcome to Python.org. In: Python.org. https://www.python.org. Accessed 15 Oct 2018
Kohavi R, John GH (1997) Wrappers for feature subset selection. Artif Intell 97:273–324
Breiman L (2001) Random Forests. Mach Learn 45:5–32
Bergstra J, Yamins D, Cox DD (2013) Hyperopt: A python library for optimizing the hyperparameters of machine learning algorithms. In: Proceedings of the 12th Python in Science Conference. Citeseer, pp 13–20
Strawderman WE (1989) Statistical Analysis with Missing Data (Roderick J. A. Little and Donald B. Rubin). SIAM Review 31:348–349
Ganeshan B, Miles KA (2013) Quantifying tumour heterogeneity with CT. Cancer Imaging 13:140–149
Cozzi L, Dinapoli N, Fogliata A, et al (2017) Radiomics based analysis to predict local control and survival in hepatocellular carcinoma patients treated with volumetric modulated arc therapy. BMC Cancer 17:829
Blanc-Durand P, Van Der Gucht A, Jreige M, et al (2018) Signature of survival: a F-FDG PET based whole-liver radiomic analysis predicts survival after Y-TARE for hepatocellular carcinoma. Oncotarget 9:4549–4558
Kim J, Choi SJ, Lee S-H, et al (2018) Predicting Survival Using Pretreatment CT for Patients With Hepatocellular Carcinoma Treated With Transarterial Chemoembolization: Comparison of Models Using Radiomics. AJR Am J Roentgenol 211:1026–1034
Hui TCH, Chuah TK, Low HM, Tan CH (2018) Predicting early recurrence of hepatocellular carcinoma with texture analysis of preoperative MRI: a radiomics study. Clin Radiol. https://doi.org/10.1016/j.crad.2018.07.109
Zhou Y, He L, Huang Y, et al (2017) CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY) 42:1695–1704
Akai H, Yasaka K, Kunimatsu A, et al (2018) Predicting prognosis of resected hepatocellular carcinoma by radiomics analysis with random survival forest. Diagn Interv Imaging 99:643–651
Li Y, Eresen A, Shangguan J, et al (2019) Establishment of a new non-invasive imaging prediction model for liver metastasis in colon cancer. Am J Cancer Res 9:2482–2492
Li M, Li X, Guo Y, et al (2020) Development and assessment of an individualized nomogram to predict colorectal cancer liver metastases. Quant Imaging Med Surg 10:397–414
Liang M, Cai Z, Zhang H, et al (2019) Machine Learning-based Analysis of Rectal Cancer MRI Radiomics for Prediction of Metachronous Liver Metastasis. Acad Radiol 26:1495–1504
Kumar V, Gu Y, Basu S, et al (2012) Radiomics: the process and the challenges. Magn Reson Imaging 30:1234–1248
Parmar C, Grossmann P, Bussink J, et al (2015) Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 5:13087
Zhang B, He X, Ouyang F, et al (2017) Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett 403:21–27
Hawkins S, Wang H, Liu Y, et al (2016) Predicting Malignant Nodules from Screening CT Scans. J Thorac Oncol 11:2120–2128
Ma J, Wang Q, Ren Y, et al (2016) Automatic lung nodule classification with radiomics approach. In: Medical Imaging 2016: PACS and Imaging Informatics: Next Generation and Innovations
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Taghavi, M., Trebeschi, S., Simões, R. et al. Machine learning-based analysis of CT radiomics model for prediction of colorectal metachronous liver metastases. Abdom Radiol 46, 249–256 (2021). https://doi.org/10.1007/s00261-020-02624-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-020-02624-1