Abstract
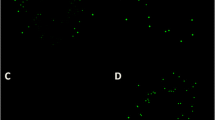
Fluorescent proteins are widely used for cell and protein tracking. Most of these proteins show a high signal and need the presence of oxygen to emit fluorescence. Among them, the fluorescent protein mCherry stands up because of its bright signal and fast maturation. Furthermore, the anaerobic cyan-green fluorescent protein Evoglow-Pp1 allows fluorescent detection under anaerobic conditions. In this work, we modified the pNZ:TuR.aFP plasmid, which harbors the gene encoding Evoglow-Pp1 and the promoter of elongation factor Tu from Limosilactobacillus reuteri CECT925, to obtain a plasmid containing the mrfp gene encoding the monomeric mCherry (pNZ:TuR.mCherry). Moreover, both genes were cloned together (pNZ:TuR.aFP.mCherry) developing a chimeric protein; and with a stop codon between them (pNZ:TuR.aFP.STOP.mCherry) resulting in the expression of both Evoglow-Pp1 and mCherry proteins separately under the influence of the same promoter. Lactococcus lactis, Lacticaseibacillus casei, Lactiplantibacillus plantarum, Limosilactobacillus fermentum, Lacticaseibacillus rhamnosus, and L. reuteri strains were transformed with the previously mentioned plasmids, showing an excellent red (pNZ:TuR.mCherry), green (pNZ:TuR.aFP), and red combined with green (pNZ:TuR.aFP.mCherry and pNZ:TuR.aFP.STOP.mCherry) fluorescence signal. Both fluorescence emissions were stable in strains transformed with pNZ:TuR.aFP.STOP.mCherry, while differences in the red or green fluorescence emission were observed in some of the strains harboring pNZ:TuR.aFP.mCherry. Moreover, these plasmids allowed strains differentiation in a complex environment, such as fecal microbiota. Hence, we present the plasmid pNZ:TuR.aFP.STOP.mCherry as a useful tool for the labeling of lactobacilli strains, which would be functional under anoxic conditions, thanks to Evoglow-Pp1, while having the high brightness and good photostability of mCherry.
Key points
• LAB transformed with pNZ:TuR.mCherry expressed the red fluorescent protein mCherry.
• LAB transformed with pNZ:TuR.aFP.mCherry developed a fusion of both proteins Evoglow-Pp1 and mCherry.
• LAB with pNZ:TuR.aFP.STOP.mCherry expressed both fluorescent proteins separately.







Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Balleza E, Kim J, Cluzel P (2018) Systematic characterization of maturation time of fluorescent proteins in living cells. Nat Methods 15:47–51
Baird GS, Zacharias DA, Tsien RY (2000) Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA 97(22):11984–11989
Baranyi J, Roberts TA (1994) A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol 23(3–4):277–294
Chudakov DM, Lukyanov S, Lukyanov KA (2005) Fluorescent proteins as a tool kit for in vivo imaging. Trends Biotechnol 23(12):605–613
Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY (1995) Understanding, improving and using green fluorescent proteins. Trends Biochem Sci 20(11):448–455
Drepper T, Eggert T, Circolone F, Heck A, Krau U, Guter K, Wendorff M, Gärtner W, Jaeger K-E (2007) Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol 25:443–445
Ernst JF, Tielker D (2009) Responses to hypoxia in fungal pathogens. Cell Microbiol 11(2):183–190
Gasson MJ (1983) Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154(1):1–9
Gaya P, Peirotén A, Landete JM (2020) Expression of a β-glucosidase in bacteria with biotechnological interest confers them the ability to deglycosylate lignans and flavonoids in vegetal foods. Appl Microbiol Biotechnol 104(11):4903–4913
Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ (2003) Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA 100(4):1990–1995
Landete JM, Arqués JL, Peirotén A, Langa S, Medina M (2014a) An improved method for the electrotransformation of lactic acid bacteria: a comparative survey. J Microbiol Meth 105:130–133
Landete JM, Langa J, Revilla C, Margolles A, Medina M, Arqués J (2015) Use of anaerobic green fluorescent protein versus green fluorescent protein as reporter in lactic acid bacteria. Appl Microbiol Biotechnol 99(16):6865–6877
Landete JM, Medina M, Arqués JL (2016) Fluorescent reporter systems for tracking probiotic lactic acid bacteria and bifidobacteria. World J Microbiol Biotechnol 32(7):119
Landete JM, Peirotén A, Rodríguez E, Margolles A, Medina M, Arqués JL (2014b) Anaerobic green fluorescent protein as a marker of Bifidobacterium strains. Int J Food Microbiol 175:6–13
Mazé A, Boël G, Zúñiga M, Bourand A, Loux V, Yebra MJ, Monedero V, Correia K, Jacques N, Beaufils S, Poncet S, Joyet P, Milohanic E, Casaregola S, Auffray Y, Pérez-Martínez G, Gibrat JF, Zagorec M, Francke C, Hartke A, Deutscher J (2010) Complete genome sequence of the probiotic Lactobacillus casei strain BL23. J Bacteriol 192(10):2647–2648
Mohedano ML, Hernández-Recio S, Yépez A, Requena T, Martínez-Cuesta MC, Peláez C, Russo P, LeBlanc JG, Spano G, Aznar R, López P (2019) Real-time detection of riboflavin production by Lactobacillus plantarum strains and tracking of their gastrointestinal survival and functionality in vitro and in vivo using mCherry labeling. Front Microbiol 10:1748
Mozzi F, Raya RR, Vignolo GM (2015) Biotechnology of lactic acid bacteria: novel applications, 2nd edn. Whiley-Blackwell, West Sussex
Müller-Taubenberger A, Anderson KI (2007) Recent advances using green and red fluorescent protein variants. Appl Microbiol Biotechnol 77(1):1–12
Peirotén A, Gaya P, Landete JM (2020) Application of recombinant lactic acid bacteria and bifidobacteria able to enrich soy beverage in dihydrodaidzein and dihydrogenistein. Food Res Int 134:109257
Rodríguez E, Arqués JL, Rodríguez R, Peirotén A, Landete JM, Medina M (2012) Antimicrobial properties of probiotic strains isolated from breast-fed infants. J Funct Foods 4(2):542–551
Russo P, Iturria I, Mohedano ML, Caggianiello G, Rainieri S, Fiocco F, Pardo MA, López P, Spano G (2015) Zebrafish gut colonization by mCherry-labelled lactic acid bacteria. Appl Microbiol Biotechnol 99(8):3479–3490
Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572
Tauer C, Heinl S, Egger E, Heiss S, Grabherr R (2014) Tuning constitutive recombinant gene expression in Lactobacillus plantarum. Microb Cell Fact 13:150
Tsien RY (1998) The green fluorescent protein. Annu Rev Biochem 67:509–544
Acknowledgements
We would like to thank undergraduate student Clara Sebastian for her assistance with fluorescence microscopy. pRCR12 plasmid harboring mrfp gene was kindly provided by Paloma López (CIB, Madrid, Spain).
Funding
This work was supported by RTA2017-00002–00-00 project from the Ministry of Economy and Competitiveness (Spain).
Author information
Authors and Affiliations
Contributions
JML conceived and designed research. JML, AP, and SL conducted experiments. JML, AP and SL analyzed the data. JML, SL, AP, and JLA interpreted the data and wrote the manuscript. All authors read and approved the submitted manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Langa, S., Peirotén, Á., Arqués, J.L. et al. Evoglow-Pp1 and mCherry proteins: a dual fluorescent labeling system for lactic acid bacteria. Appl Microbiol Biotechnol 105, 7367–7378 (2021). https://doi.org/10.1007/s00253-021-11537-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11537-y