Abstract
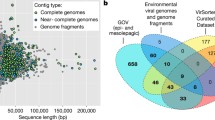
Compared to free-living viruses (< 0.22 m) in the ocean, planktonic viruses in the “cellular fraction” (0.22 ~ 3.0 μm) are now far less well understood, and the differences between them remain largely unexplored. Here, we revealed that even in the same seawater samples, the “cellular fraction” comprised significantly distinct virus communities from the free virioplankton, with only 13.87% overlap in viral contigs at the species level. Compared to the viral genomes deposited in NCBI RefSeq database, 99% of the assembled viral genomes in the “cellular fraction” represented novel genera. Notably, the assembled (near-) complete viral genomes within the “cellular fraction” were significantly larger than that in the “viral fraction,” and the “cellular fraction” contained three times more species of giant viruses or jumbo phages with genomes > 200 kb than the “viral fraction.” The longest complete genomes of jumbo phage (~ 252 kb) and giant virus (~ 716 kb) were both detected only in the “cellular fraction.” Moreover, a relatively higher proportion of proviruses were predicted within the “cellular fraction” than “viral fraction.” Besides the substantial divergence in viral community structure, the different fractions also contained their unique viral auxiliary metabolic genes; e.g., those potentially participating in inorganic carbon fixation in deep sea were detected only in the “cellular-fraction” viromes. In addition, there was a considerable divergence in the community structure of both “cellular fraction” and “viral fraction” viromes between the surface and deep-sea habitats, suggesting that they might have similar environmental adaptation properties. The findings deepen our understanding of the complexity of viral community structure and function in the ocean.






Similar content being viewed by others
Data Availability
The CVC and VVC datasets reported in this study have been deposited in the Genome Warehouse in the National Genomics Data Center [104], Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under BioProject accession number PRJCA002898 and BioSample accession number SAMC231958 and SAMC231959, respectively. The CVG and VVG datasets have been deposited under BioSample accession number SAMC195331 and SAMC195332, respectively. The C-vMAGs and V-vMAGs datasets have been deposited under BioSample accession number SAMC1000078 and SAMC1000079, respectively. They are publicly accessible at https://bigd.big.ac.cn/gwh. Raw data files and R codes for statistical analyses have been deposited in the Figshare: https://doi.org/10.6084/m9.figshare.c.5984017.v4.
References
Dion MB, Oechslin F, Moineau S (2020) Phage diversity, genomics and phylogeny. Nat Rev Microbiol 18:125–138. https://doi.org/10.1038/s41579-019-0311-5
Hurwitz BL, Sullivan MB (2013) The Pacific Ocean Virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS ONE 8:e57355. https://doi.org/10.1371/journal.pone.0057355
Brum JR, Ignacio-Espinoza JC, Roux S et al (2015) Patterns and ecological drivers of ocean viral communities. Science 348:1261498. https://doi.org/10.1126/science.1261498
Gregory AC, Zayed AA, Conceicao-Neto N et al (2019) Marine DNA viral macro- and microdiversity from pole to pole. Cell 177:1109–1123. https://doi.org/10.1016/j.cell.2019.03.040
Rontani JF, Zabeti N, Wakeham SG (2011) Degradation of particulate organic matter in the equatorial Pacific Ocean: biotic or abiotic? Limnol Oceanogr 56:333–349. https://doi.org/10.4319/lo.2011.56.1.0333
Yamada Y, Guillemette R, Baudoux AC et al (2020) Viral attachment to biotic and abiotic surfaces in seawater. Appl Environ Microbiol 86:e01687-e1619. https://doi.org/10.1128/AEM.01687-19
Brussow H (2020) Huge bacteriophages: bridging the gap? Environ Microbiol 22:1965–1970. https://doi.org/10.1111/1462-2920.15034
Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA et al (2016) Uncovering Earth’s virome. Nature 536:425–430. https://doi.org/10.1038/nature19094
Coutinho FH, Gregoracci GB, Walter JM et al (2018) Metagenomics sheds light on the ecology of marine microbes and their viruses. Trends Microbiol 26:955–965. https://doi.org/10.1016/j.tim.2018.05.015
Lopez-Perez M, Haro-Moreno JM, de la Torre JR, Rodriguez-Valera F (2019) Novel Caudovirales associated with Marine Group I Thaumarchaeota assembled from metagenomes. Environ Microbiol 21:1980–1988. https://doi.org/10.1111/1462-2920.14462
Zheng XW, Liu W, Dai X et al (2021) Extraordinary diversity of viruses in deep-sea sediments as revealed by metagenomics without prior virion separation. Environ Microbiol 23:728–743. https://doi.org/10.1111/1462-2920.15154
Flores-Uribe J, Philosof A, Sharon I et al (2019) A novel uncultured marine cyanophage lineage with lysogenic potential linked to a putative marine Synechococcus ‘relic’ prophage. Environ Microbiol Rep 11:598–604. https://doi.org/10.1111/1758-2229.12773
Palermo CN, Shea DW, Short SM (2021) Analysis of different size fractions provides a more complete perspective of viral diversity in a freshwater embayment. Appl Environ Microbiol 87:e00197-e121. https://doi.org/10.1128/AEM.00197-21
Lopez-Perez M, Haro-Moreno JM, Gonzalez-Serrano R et al (2017) Genome diversity of marine phages recovered from Mediterranean metagenomes: Size matters. PLoS Genet 13:e1007018. https://doi.org/10.1371/journal.pgen.1007018
Roux S, Enault F, Hurwitz BL, Sullivan MB (2015) VirSorter: mining viral signal from microbial genomic data. PeerJ 3:e985. https://doi.org/10.7717/peerj.985
Nayfach S, Camargo AP, Schulz F et al (2021) CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol 39:578–585. https://doi.org/10.1038/s41587-020-00774-7
Kieft K, Zhou ZC, Anantharaman K (2020) VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 8:1–23. https://doi.org/10.1186/s40168-020-00867-0
Wang Z, Zhao J, Wang L et al (2019) A novel benthic phage infecting Shewanella with strong replication ability. Viruses 11:1081. https://doi.org/10.3390/v11111081
Chen J, Li HM, Zhang ZH et al (2020) DOC dynamics and bacterial community succession during long-term degradation of Ulva prolifera and their implications for the legacy effect of green tides on refractory DOC pool in seawater. Water Res 185:116268. https://doi.org/10.1016/j.watres.2020.116268
Forterre P, Soler N, Krupovic M et al (2013) Fake virus particles generated by fluorescence microscopy. Trends Microbiol 21:1–5. https://doi.org/10.1016/j.tim.2012.10.005
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. https://doi.org/10.1101/gr.213959.116
Li DH, Liu CM, Luo RB et al (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. https://doi.org/10.1093/bioinformatics/btv033
Gao SM, Schippers A, Chen N et al (2020) Depth-related variability in viral communities in highly stratified sulfidic mine tailings. Microbiome 8:1–13. https://doi.org/10.1186/s40168-020-00848-3
Fu L, Niu B, Zhu Z et al (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Roux S, Brum JR, Dutilh BE et al (2016) Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537:689–693. https://doi.org/10.1038/nature19366
Shen W, Le S, Li Y, Hu FQ (2016) SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 11:e0163962. https://doi.org/10.1371/journal.pone.0163962
Roux S, Adriaenssens EM, Dutilh BE et al (2019) Minimum information about an uncultivated virus genome (MIUViG). Nat Biotechnol 37:29–37. https://doi.org/10.1038/nbt.4306
Jian H, Yi Y, Wang J et al (2021) Diversity and distribution of viruses inhabiting the deepest ocean on Earth. ISME J 15:3094–3110. https://doi.org/10.1038/s41396-021-00994-y
Li Z, Pan D, Wei G et al (2021) Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. ISME J 15:2366–2378. https://doi.org/10.1038/s41396-021-00932-y
Liao H, Li H, Duan CS et al (2022) Response of soil viral communities to land use changes. Nat Commun 13:6027. https://doi.org/10.1038/s41467-022-33771-2
Weinheimer AR, Aylward FO (2022) Infection strategy and biogeography distinguish cosmopolitan groups of marine jumbo bacteriophages. ISME J 16:1657–1667. https://doi.org/10.1038/s41396-022-01214-x
Roux S, Emerson JB, Eloe-Fadrosh EA, Sullivan MB (2017) Benchmarking viromics: an in silico evaluation of metagenome-enabled estimates of viral community composition and diversity. PeerJ 5:e3817. https://doi.org/10.7717/peerj.3817
Jang HB, Bolduc B, Zablocki O et al (2019) Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol 37:632–639. https://doi.org/10.1038/s41587-019-0100-8
Pratama AA, Bolduc B, Zayed AA et al (2021) Expanding standards in viromics: in silico evaluation of dsDNA viral genome identification, classification, and auxiliary metabolic gene curation. PeerJ 9:e11447. https://doi.org/10.7717/peerj.11447
Pons JC, Paez-Espino D, Riera G et al (2021) VPF-Class: taxonomic assignment and host prediction of uncultivated viruses based on viral protein families. Bioinformatics 37:1805–1813. https://doi.org/10.1093/bioinformatics/btab026
Hyatt D, Chen GL, LoCascio PF et al (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:1–11. https://doi.org/10.1186/1471-2105-11-119
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Roux S, Paez-Espino D, Chen IMA et al (2021) IMG/VR v3: an integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res 49:D764–D775. https://doi.org/10.1093/nar/gkaa946
Hockenberry AJ, Wilke CO (2021) BACPHLIP: predicting bacteriophage lifestyle from conserved protein domains. PeerJ 9:e11396. https://doi.org/10.7717/peerj.11396
Huerta-Cepas J, Szklarczyk D, Heller D et al (2019) eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. https://doi.org/10.1093/nar/gky1085
Shaffer M, Borton MA, McGivern BB et al (2020) DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res 48:8883–8900. https://doi.org/10.1093/nar/gkaa621
Lu SN, Wang JY, Chitsaz F et al (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268. https://doi.org/10.1093/nar/gkz991
Kelley LA, Mezulis S, Yates CM et al (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. https://doi.org/10.1038/nprot.2015.053
Nishimura Y, Yoshida T, Kuronishi M et al (2017) ViPTree: the viral proteomic tree server. Bioinformatics 33:2379–2380. https://doi.org/10.1093/bioinformatics/btx157
Adriaenssens EM, Cowan DA (2014) Using signature genes as tools to assess environmental viral ecology and diversity. Appl Environ Microbiol 80:4470–4480. https://doi.org/10.1128/AEM.00878-14
Jin M, Guo X, Zhang R et al (2019) Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome 7:1–15. https://doi.org/10.1186/s40168-019-0675-9
Zhao JL, Jing HM, Wang ZM et al (2022) Novel viral communities potentially assisting in carbon, nitrogen, and sulfur metabolism in the upper slope sediments of Mariana Trench. mSystems 7: e01358–01321. doi: https://doi.org/10.1128/msystems.01358-21
Lu J, Yang SX, Zhang XD et al (2022) Metagenomic analysis of viral community in the Yangtze River expands known eukaryotic and prokaryotic virus diversity in freshwater. Virol Sin 37:60–69. https://doi.org/10.1016/j.virs.2022.01.003
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. https://doi.org/10.1093/nar/gki198
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Price MN, Dehal PS, Arkin AP (2010) FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. https://doi.org/10.1371/journal.pone.0009490
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. https://doi.org/10.1093/nar/gkab301
Uritskiy GV, DiRuggiero J, Taylor J (2018) MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6:1–13. https://doi.org/10.1186/s40168-018-0541-1
Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH (2020) GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. https://doi.org/10.1093/bioinformatics/btz848
Rho M, Wu YW, Tang HX et al (2012) Diverse CRISPRs evolving in human microbiomes. PLoS Genet 8:e1002441. https://doi.org/10.1371/journal.pgen.1002441
Huntemann M, Ivanova NN, Mavromatis K et al (2016) The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP vol 4). Stand Genomic Sci 10:1–6. https://doi.org/10.1186/s40793-016-0148-8
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. https://doi.org/10.1093/nar/gki366
Roux S, Camargo AP, Coutinho FH et al (2022) iPHoP: an integrated machine-learning framework to maximize host prediction for metagenome-assembled virus genomes. bioRxiv: 2022.2007.2028.501908. https://doi.org/10.1101/2022.07.28.501908
Garcia-Lopez R, Vazquez-Castellanos JF, Moya A (2015) Fragmentation and coverage variation in viral metagenome assemblies, and their effect in diversity calculations. Front Bioeng Biotechnol 3:141. https://doi.org/10.3389/fbioe.2015.00141
Johansen J, Plichta DR, Nissen JN et al (2022) Genome binning of viral entities from bulk metagenomics data. Nat Commun 13:965. https://doi.org/10.1038/s41467-022-28581-5
Olm MR, Brown CT, Brooks B, Banfield JF (2017) dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J 11:2864–2868. https://doi.org/10.1038/ismej.2017.126
Willis A, Bunge J (2015) Estimating diversity via frequency ratios. Biometrics 71:1042–1049. https://doi.org/10.1111/biom.12332
Oksanen J, Blanchet F, Friendly M et al (2018) vegan: community ecology package. R package version 2.5–2.
Wickham H (2016) ggplot2 – elegant graphics for data analysis, 2nd edn. Springer-Verlag, New York
Chen T, Liu Y-X, Huang L (2022) ImageGP: an easy-to-use data visualization web server for scientific researchers. iMeta 1: e5. https://doi.org/10.1002/imt2.5
Nishimura Y, Watai H, Honda T et al (2017) Environmental viral genomes shed new light on virus-host interactions in the ocean. mSphere 2: e00359–00316. https://doi.org/10.1128/mSphere.00359-16
Mihara T, Nishimura Y, Shimizu Y et al (2016) Linking virus genomes with host taxonomy. Viruses 8:66. https://doi.org/10.3390/v8030066
Yuan YH, Gao MY (2017) Jumbo bacteriophages: an overview. Front Microbiol 8:403. https://doi.org/10.3389/fmicb.2017.00403
Breitbart M, Bonnain C, Malki K, Sawaya NA (2018) Phage puppet masters of the marine microbial realm. Nat Microbiol 3:754–766. https://doi.org/10.1038/s41564-018-0166-y
Needham DM, Yoshizawa S, Hosaka T et al (2019) A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators. Proc Natl Acad Sci USA 116:20574–20583. https://doi.org/10.1073/pnas.1907517116
Zhang W, Zhou J, Liu T et al (2015) Four novel algal virus genomes discovered from Yellowstone Lake metagenomes. Sci Rep 5:15131. https://doi.org/10.1038/srep15131
Moniruzzaman M, Martinez-Gutierrez CA, Weinheimer AR, Aylward FO (2020) Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Nat Commun 11:1710. https://doi.org/10.1038/s41467-020-15507-2
Bowers RM, Kyrpides NC, Stepanauskas R et al (2017) Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. https://doi.org/10.1038/nbt.3893
Gill JJ, Berry JD, Russell WK et al (2012) The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genomics 13:1–21. https://doi.org/10.1186/1471-2164-13-542
Schvarcz CR, Steward GF (2018) A giant virus infecting green algae encodes key fermentation genes. Virology 518:423–433. https://doi.org/10.1016/j.virol.2018.03.010
Zaczek-Moczydłowska MA, Young GK, Trudgett J et al (2020) Genomic characterization, formulation and efficacy in planta of a Siphoviridae and Podoviridae protection cocktail against the bacterial plant pathogens Pectobacterium spp. Viruses 12:150. https://doi.org/10.3390/v12020150
Gao C, Liang Y, Jiang Y et al (2022) Virioplankton assemblages from challenger deep, the deepest place in the oceans. iScience 25:104680. https://doi.org/10.1016/j.isci.2022.104680
Jin M, Cai L, Ma R et al (2020) Prevalence of temperate viruses in deep South China Sea and western Pacific Ocean. Deep-Sea Res Pt I. https://doi.org/10.1016/j.dsr.2020.103403
Gong Z, Liang Y, Wang M et al (2018) Viral diversity and its relationship with environmental factors at the surface and deep sea of Prydz Bay. Antarctica Front Microbiol 9:2981. https://doi.org/10.3389/fmicb.2018.02981
Cook R, Brown N, Redgwell T et al (2021) INfrastructure for a PHAge REference Database: identification of large-scale biases in the current collection of cultured phage genomes. PHAGE 2:214–223. https://doi.org/10.1089/phage.2021.0007
Ceyssens P-J, Minakhin L, Van den Bossche A et al (2014) Development of giant bacteriophage ϕKZ is independent of the host transcription apparatus. J Virol 88:10501–10510. https://doi.org/10.1128/JVI.01347-14
Van den Bossche A, Hardwick SW, Ceyssens P-J et al (2016) Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome. Elife 5:e16413. https://doi.org/10.7554/eLife.164131
Al-Shayeb B, Sachdeva R, Chen LX et al (2020) Clades of huge phages from across Earth’s ecosystems. Nature 578:425–431. https://doi.org/10.1038/s41586-020-2007-4
Brandes N, Linial M (2019) Giant viruses-big surprises. Viruses 11:404. https://doi.org/10.3390/v11050404
Schulz F, Abergel C, Woyke T (2022) Giant virus biology and diversity in the era of genome-resolved metagenomics. Nat Rev Microbiol 20:721–736. https://doi.org/10.1038/s41579-022-00754-5
Ogata H, Ray J, Toyoda K et al (2011) Two new subfamilies of DNA mismatch repair proteins (MutS) specifically abundant in the marine environment. ISME J 5:1143–1151. https://doi.org/10.1038/ismej.2010.210
Hingamp P, Grimsley N, Acinas SG et al (2013) Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J 7:1678–1695. https://doi.org/10.1038/ismej.2013.59
Raoult D, Audic S, Robert C et al (2004) The 1.2-megabase genome sequence of Mimivirus. Science 306:1344–1350. https://doi.org/10.1126/science.1101485
Endo H, Blanc-Mathieu R, Li YZ et al (2020) Biogeography of marine giant viruses reveals their interplay with eukaryotes and ecological functions. Nat Ecol Evol 4:1639–1649. https://doi.org/10.1038/s41559-020-01288-w
Pound HL, Gann ER, Tang XM et al (2020) The “neglected viruses” of Taihu: abundant transcripts for viruses infecting eukaryotes and their potential role in phytoplankton succession. Front Microbiol 11:338. https://doi.org/10.3389/fmicb.2020.00338
Kwon J, Kim SG, Kim HJ et al (2021) Isolation and characterization of Salmonella jumbo-phage pSal-SNUABM-04. Viruses 13:27. https://doi.org/10.3390/v13010027
Yamada T, Onimatsu H, Van Etten JL (2006) Chlorella viruses. Adv Virus Res. Academic Press, pp. 293–336
Payet JP, Suttle CA (2013) To kill or not to kill: The balance between lytic and lysogenic viral infection is driven by trophic status. Limnol Oceanogr 58:465–474. https://doi.org/10.4319/lo.2013.58.2.0465
Silveira CB, Luque A, Rohwer F (2021) The landscape of lysogeny across microbial community density, diversity and energetics. Environ Microbiol 23:4098–4111. https://doi.org/10.1111/1462-2920.15640
Tuttle MJ, Buchan A (2020) Lysogeny in the oceans: lessons from cultivated model systems and a reanalysis of its prevalence. Environ Microbiol 22:4919–4933. https://doi.org/10.1111/1462-2920.15233
Luo E, Eppley JM, Romano AE et al (2020) Double-stranded DNA virioplankton dynamics and reproductive strategies in the oligotrophic open ocean water column. ISME J 14:1304–1315. https://doi.org/10.1038/s41396-020-0604-8
Bongiorni L, Magagnini M, Armeni M et al (2005) Viral production, decay rates, and life strategies along a trophic gradient in the north Adriatic sea. Appl Environ Microbiol 71:6644–6650. https://doi.org/10.1128/AEM.71.11.6644-6650.2005
Lipson SM, Stotzky G (1984) Effect of proteins on reovirus adsorption to clay-minerals. Appl Environ Microbiol 48:525–530. https://doi.org/10.1128/AEM.48.3.525-530.1984
Wu S, Zhou L, Zhou YF et al (2020) Diverse and unique viruses discovered in the surface water of the East China Sea. BMC Genomics 21:1–15. https://doi.org/10.1186/s12864-020-06861-y
Yu AL, Xie Y, Pan XW et al (2020) Photosynthetic phosphoribulokinase structures: enzymatic mechanisms and the redox regulation of the Calvin-Benson-Bassham Cycle. Plant Cell 32:1556–1573. https://doi.org/10.1105/tpc.19.00642
Kanao T, Kawamura M, Fukui T et al (2002) Characterization of isocitrate dehydrogenase from the green sulfur bacterium Chlorobium limicola - a carbon dioxide-fixing enzyme in the reductive tricarboxylic acid cycle. Eur J Biochem 269:1926–1931. https://doi.org/10.1046/j.1432-1033.2002.02849.x
Hugler M, Sievert SM (2011) Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann Rev Mar Sci 3:261–289. https://doi.org/10.1146/annurev-marine-120709-142712
Mizuno Carolina M, Ghai R, Saghaï A et al (2016) Genomes of abundant and widespread viruses from the deep ocean. mBio 7: e00805–00816. https://doi.org/10.1128/mBio.00805-16
Zhang Z, Zhao WM, Xiao JF et al (2020) Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res 48:D24–D33. https://doi.org/10.1093/nar/gkz913
Funding
This work was funded by the National Natural Science Foundation of China (No. 41876174, 42206124, 42106107, 42006093); the Senior User Project of RV KEXUE (KEXUE2019GZ03) supported by the Center for Ocean Mega-Science, Chinese Academy of Sciences; and the open research cruise NORC2017-05 supported by an NSFC Ship-time Sharing Project.
Author information
Authors and Affiliations
Contributions
Y.Z. and N.J. designed the experiment. J.Z. and Z.W. conducted the experiments and performed the metagenomic analysis. C.L., T.S., and Y.L. collected the samples. J.Z., Z.W., and Y.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Consent for Publication
All the authors give their consent to allow this manuscript to be published by the Journal of Microbial Ecology.
Conflict of Interest
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
248_2022_2167_MOESM1_ESM.pdf
Supplementary file1 Fig. S1: Workflow of metagenomic sampling, sequencing, assembly, and creation of the South China Sea viral genome datasets. Fig. S2: Abundance profiles of free virus-like particles and prokaryotes in the seawater. (a) The abundance of virus-like particles and prokaryotes in all the samples. The virus-to-prokaryote ratio (VPR) of each sample was calculated by dividing the abundance of free virus-like particles by the abundance of prokaryotes. The error bars represent the standard deviation of three measurements. (b) The correlation between prokaryotic abundance and viral-like particle (VLP) abundance in all the samples. Fig. S3: Distribution of size and GC content (%) of CVCs/VVCs (a) and CVGs/ VVGs (b). Fig. S4: Phylogenetic analysis of the CVGs obtained from the ‘cellular-fraction’ metagenomes and reference viral genomes. Branches in red indicate the viral genomes from this study, whereas those in black indicate reference viral genomes retrieved from the NCBI RefSeq Virus database. The rings outside the tree represent (from inside to outside) the CVGs, the virus family, and the host group. Fig. S5: Phylogenetic analysis of the VVGs obtained from the ‘viral-fraction’ metagenomesand reference viral genomes. Branches in red indicate the viral genomes from this study, whereas those in black indicate the reference viral genomes retrieved from the NCBI RefSeq Virus database. The rings outside the tree represent (from inside to outside) the VVGs, the virus family, and the host group. Fig. S6: Genome map of CVG_41. The rings (from inside to outside) represent GC Skew (Ring 1), GC content (Ring 2), the direction of transcription (Ring 3), and gene features (i.e., functional genes, hypothetical protein, and unknown function) (Ring 4). Fig. S7: Annotation of the genes encoded by viruses in the CVC (a) and VVC (b) datasets against the eggNOG 5.0 database. Colors in pie charts represent the number of genes; red as the annotated genes, and blue as unknown. Fig. S8: Percentage of viral contigs having predicted hosts in the ‘cellular fraction’ and ‘viral fraction’ (pie charts), and predicted host composition grouped by phylum (class for Proteobacteria) in the two fractions (bar plots). Fig. S9: Proteomic tree representing the proteome-wide similarity relationships between the longest viral MAGs (C-S3C4721) and 112 reference eukaryotic viral genomes. The inner ring represents the virus family, and the outer ring represents the host group. (PDF 13361 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, J., Wang, Z., Li, C. et al. Significant Differences in Planktonic Virus Communities Between “Cellular Fraction” (0.22 ~ 3.0 µm) and “Viral Fraction” (< 0.22 μm) in the Ocean. Microb Ecol 86, 825–842 (2023). https://doi.org/10.1007/s00248-022-02167-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-022-02167-6