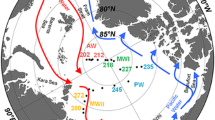
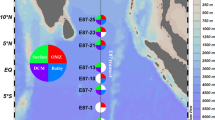
Abstract
The Arctic Ocean is facing rapid environmental changes with cascading effects on the entire Arctic marine ecosystem. However, we have a limited understanding of the consequences such changes have on bacteria and archaea (prokaryotes) at the base of the marine food web. In this study, we show how the prokaryotic rare biosphere behaves over a range of highly heterogeneous environmental conditions using 16S rRNA gene reads from amplicon and metagenome sequencing data from seawater samples collected during the Norwegian young sea ICE expedition between late winter and early summer. The prokaryotic rare biosphere was analyzed using different approaches: amplicon sequence variants and operational taxonomic units from the 16S rRNA gene amplicons and operational taxonomic units from the 16S rRNA genes of the metagenomes. We found that prokaryotic rare biosphere communities are specific to certain water masses, and that the majority of the rare taxa identified were always rare and disappeared in at least one sample under changing conditions, suggesting their high sensitivity to environmental heterogeneity. In addition, our methodological comparison revealed a good performance of 16S rRNA gene amplicon sequencing in describing rare biosphere patterns, while the metagenome-derived data were better to capture a significant diversity of so-far uncultivated rare taxa. Our analysis on the dynamics of the rare prokaryotic biosphere, by combining different methodological approaches, improves the description of the types of rarity predicted from Community Assembly theory in the Arctic Ocean.




Similar content being viewed by others
Data availability
The raw sequences used for 16S rRNA gene analysis and metagenomes are publically available at the European Nucleotide Archive (ENA) under the accession numbers PRJEB21950 and PRJEB15043. The sample vs. 16S rRNA gene–derived OTUs table is available in the MGnify platform (Study: MGYS00001922). The sample versus metagenomic-derived OTUs (mOTU) table is available at the MGnify platform (Study: MGYS00001869).
References
Meyer A, Sundfjord A, Fer I, Provost C, Villacieros Robineau N, Koenig Z, Onarheim IH, Smedsrud LH, Duarte P, Dodd PA, Graham RM, Schmidtko S, Kauko HM (2017) Winter to summer oceanographic observations in the Arctic Ocean north of Svalbard. J Geophys Res Oceans 122:6218–6237. https://doi.org/10.1002/2016JC012391
Müller O, Wilson B, Paulsen ML, Rumińska A, Armo HR, Bratbak G, Øvreås L (2018) Spatiotemporal dynamics of ammonia-oxidizing Thaumarchaeota in distinct Arctic water masses. Front Microbiol 9:24. https://doi.org/10.3389/fmicb.2018.00024
Wilson B, Müller O, Nordmann EL, Seuthe L, Bratbak G, Øvreås L (2017) Changes in marine prokaryote composition with season and depth over an Arctic polar year. Front Mar Sci 4:95. https://doi.org/10.3389/fmars.2017.00095
de Sousa AGG, Tomasino MP, Duarte P, Fernández-Méndez M, Assmy P, Ribeiro H, Surkont J, Leite RB, Pereira-Leal JB, Torgo L, Magalhães C (2019) Diversity and composition of pelagic prokaryotic and protist communities in a thin Arctic Sea-Ice regime. Microb Ecol 78:388–408. https://doi.org/10.1007/s00248-018-01314-2
Galand PE, Casamayor EO, Kirchman DL, Lovejoy C (2009) Ecology of the rare microbial biosphere of the Arctic Ocean. PNAS 106:22427–22432. https://doi.org/10.1073/pnas.0908284106
Kirchman DL, Cottrell MT, Lovejoy C (2010) The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol 12:1132–1143. https://doi.org/10.1111/j.1462-2920.2010.02154.x
Ghiglione J-F, Galand PE, Pommier T, Pedros-Alio C, Maas EW, Bakker K, Bertilson S, Kirchman DL, Lovejoy C, Yager PL, Murray AE (2012) Pole-to-pole biogeography of surface and deep marine bacterial communities. PNAS 109:17633–17638. https://doi.org/10.1073/pnas.1208160109
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. PNAS 103:12115–12120. https://doi.org/10.1073/pnas.0605127103
Pedrós-Alió C (2006) Marine microbial diversity: can it be determined? Trends Microbiol 14:257–263. https://doi.org/10.1016/j.tim.2006.04.007
Shade A, Jones SE, Caporaso JG, Handelsman J, Knight R, Fierer N, Gilbert JA (2014) Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. MBio 5:1–9. https://doi.org/10.1128/mBio.01371-14
Jia X, Dini-Andreote F, Salles JF (2018) Community assembly processes of the microbial rare biosphere. Trends Microbiol 26:738–747. https://doi.org/10.1016/j.tim.2018.02.011
Pascoal F, Costa R, Magalhães C (2021) The microbial rare biosphere: current concepts, methods and ecological principles. FEMS Microbiol Ecol 97:1. https://doi.org/10.1093/femsec/fiaa227
Vellend M, Agrawal A (2010) Conceptual synthesis in community ecology. Q Rev Biol 85:183–206. https://doi.org/10.1086/652373
Zhou J, Ning D (2017) Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:e00002–e00017. https://doi.org/10.1128/MMBR.00002-17
Jousset A, Bienhold C, Chatzinotas A, Gallien L, Gobet A, Kurm V, Küsel K, Rillig MC, Rivett DW, Salles JF, van der Heijden MGA, Youssef NH, Zhang X, Wei Z, Hol WHG (2017) Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J 11:853–862. https://doi.org/10.1038/ismej.2016.174
Fernández-Gómez B, Díez B, Polz MF, Arroyo JI, Alfaro FD, Marchandon G, Sanhueza C, Farías L, Trefault N, Marquet PA, Molina-Montenegro MA, Sylvander P, Snoeijs-Leijonmalm P (2019) Bacterial community structure in a sympagic habitat expanding with global warming: brackish ice brine at 85–90 °N. ISME J 13:316–333. https://doi.org/10.1038/s41396-018-0268-9
Baltar F, Currie K, Stuck E, Roosa S, Morales SE (2016) Oceanic fronts: transition zones for bacterioplankton community composition. Environ Microbiol Rep 8:132–138. https://doi.org/10.1111/1758-2229.12362
Gokul JK, Cameron KA, Irvine-Fynn TDL, Cook JM, Hubbard A, Stibal M, Hegarty M, Mur LAJ, Edwards A (2019) Illuminating the dynamic rare biosphere of the Greenland Ice Sheet’s Dark Zone. FEMS Microbiol Ecol 95:12. https://doi.org/10.1093/femsec/fiz177
Granskog MA, Fer I, Rinke A, Steen H (2018) Atmosphere-ice-ocean-ecosystem processes in a thinner Arctic sea ice regime: The Norwegian Young Sea ICE (N-ICE2015) Expedition. J Geophys Res Oceans 123:1586–1594. https://doi.org/10.1002/2017JC013328
Kopf A, Bicak M, Kottmann R, Schnetzer J, Kostadinov I, Lehmann K, Fernandez-Guerra A, Jeanthon C, Rahav E, Ullrich M, Wichels A, Gerdts G, Polymenakou P, Kotoulas G, Siam R, Abdallah RZ, Sonnenschein EC, Cariou T, O’Gara F, Jackson S, Orlic S, Steinke M, Busch J, Duarte B, Caçador I, Canning-Clode J, Bobrova O, Marteinsson V, Reynisson E, Loureiro CM, Luna GM, Quero GM, Löscher CR, Kremp A, DeLorenzo ME, Øvreås L, Tolman J, LaRoche J, Penna A, Frischer M, Davis T, Katherine B, Meyer CP, Ramos S, Magalhães C, Jude-Lemeilleur F, Aguirre-Macedo ML, Wang S, Poulton N, Jones S, Collin R, Fuhrman JA, Conan P, Alonso C, Stambler N, Goodwin K, Yakimov MM, Baltar F, Bodrossy L, van de Kamp J, Frampton DMF, Ostrowski M, van Ruth P, Malthouse P, Claus S, Deneudt K, Mortelmans J, Pitois S, Wallom D, Salter I, Costa R, Schroeder DC, Kandil MM, Amaral V, Biancalana F, Santana R, Pedrotti ML, Yoshida T, Ogata H, Ingleton T, Munnik K, Rodriguez-Ezpeleta N, Berteaux-Lecellier V, Wecker P, Cancio I, Vaulot D, Bienhold C, Ghazal H, Chaouni B, Essayeh S, Ettamimi S, Zaid EH, Boukhatem N, Bouali A, Chahboune R, Barrijal S, Timinouni M, el Otmani F, Bennani M, Mea M, Todorova N, Karamfilov V, ten Hoopen P, Cochrane G, L’Haridon S, Bizsel KC, Vezzi A, Lauro FM, Martin P, Jensen RM, Hinks J, Gebbels S, Rosselli R, de Pascale F, Schiavon R, dos Santos A, Villar E, Pesant S, Cataletto B, Malfatti F, Edirisinghe R, Silveira JAH, Barbier M, Turk V, Tinta T, Fuller WJ, Salihoglu I, Serakinci N, Ergoren MC, Bresnan E, Iriberri J, Nyhus PAF, Bente E, Karlsen HE, Golyshin PN, Gasol JM, Moncheva S, Dzhembekova N, Johnson Z, Sinigalliano CD, Gidley ML, Zingone A, Danovaro R, Tsiamis G, Clark MS, Costa AC, el Bour M, Martins AM, Collins RE, Ducluzeau AL, Martinez J, Costello MJ, Amaral-Zettler LA, Gilbert JA, Davies N, Field D, Glöckner FO (2015) The ocean sampling day consortium. Gigascience 4:27. https://doi.org/10.1186/s13742-015-0066-5
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS 108:4516–4522. https://doi.org/10.1073/pnas.1000080107
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. https://doi.org/10.1038/ismej.2012.8
Apprill A, McNally S, Parsons R, Weber L (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. https://doi.org/10.3354/ame01753
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. https://doi.org/10.1111/1462-2920.13023
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Mitchell AL, Almeida A, Beracochea M, Boland M, Burgin J, Cochrane G, Crusoe MR, Kale V, Potter SC, Richardson LJ, Sakharova E, Scheremetjew M, Korobeynikov A, Shlemov A, Kunyavskaya O, Lapidus A, Finn RD (2019) MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res 48:D570–D578. https://doi.org/10.1093/nar/gkz1035
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
R Core Team (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
Villanueva RAM, Chen ZJ (2019) ggplot2: elegant graphics for data analysis (2nd ed.). Meas Interdiscip Res Perspect 17:160–167. https://doi.org/10.1080/15366367.2019.1565254
Oksanen J, Guillaume Blanchet F, Friendly M et al (2018) Community ecology package. R Package Version 2:5–3 https://cran.r-project.org
Gobet A, Quince C, Ramette A (2010) Multivariate Cutoff Level Analysis (MultiCoLA) of large community data sets. Nucleic Acids Res 38:e155–e155. https://doi.org/10.1093/nar/gkq545
Pedrós-Alió C (2012) The rare bacterial biosphere. Ann Rev Mar Sci 4:449–466. https://doi.org/10.1146/annurev-marine-120710-100948
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. https://doi.org/10.1101/gr.092759.109
McMurdie PJ, Holmes S (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. https://doi.org/10.1371/journal.pcbi.1003531
Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF (2016) A new view of the tree of life. Nat Microbiol 1:16048. https://doi.org/10.1038/nmicrobiol.2016.48
Needham DM, Sachdeva R, Fuhrman JA (2017) Ecological dynamics and co-occurrence among marine phytoplankton, bacteria and myoviruses shows microdiversity matters. ISME J 11:1614–1629. https://doi.org/10.1038/ismej.2017.29
Martinez-Gutierrez CA, Aylward FO (2019) Strong purifying selection is associated with genome streamlining in epipelagic marinimicrobia. Genome Biol Evol 11:2887–2894. https://doi.org/10.1093/gbe/evz201
Bowman JS, Rasmussen S, Blom N, Deming JW, Rysgaard S, Sicheritz-Ponten T (2012) Microbial community structure of Arctic multiyear sea ice and surface seawater by 454 sequencing of the 16S RNA gene. ISME J 6:11–20. https://doi.org/10.1038/ismej.2011.76
Glassman SI, Martiny JBH (2018) Broadscale ecological patterns are robust to use of exact sequence variants versus operational taxonomic units. mSphere 3:e00148–e00118. https://doi.org/10.1128/mSphere.00148-18
Bay SK, McGeoch MA, Gillor O et al (2020) Soil bacterial communities exhibit strong biogeographic patterns at fine taxonomic resolution. mSystems 5:1–16. https://doi.org/10.1128/msystems.00540-20
Elshahed MS, Youssef NH, Spain AM, Sheik C, Najar FZ, Sukharnikov LO, Roe BA, Davis JP, Schloss PD, Bailey VL, Krumholz LR (2008) Novelty and uniqueness patterns of rare members of the soil biosphere. Appl Environ Microbiol 74:5422–5428. https://doi.org/10.1128/AEM.00410-08
Pascoal F, Magalhães C, Costa R (2020) The link between the ecology of the prokaryotic rare biosphere and its biotechnological potential. Front Microbiol 11:231. https://doi.org/10.3389/fmicb.2020.00231
Code availability
The code used for processing of raw 16S rRNA gene sequences into ASVs and the code for the diversity metrics used for all datasets (ASVs, OTUs, and mOTUs) are available in a single R script (Supplementary Data S1). Custom commands were used for the Circos figures, based on the data obtained from the R script provided.
Funding
The Portuguese Science and Technology Foundation (FCT) funded this study through the grants PTDC/CTA-AMB/30997/2017 and PTDC/CTA-AMB/4946/2020 to C.M., 2020.03139 CEECIND to C.M., and the PhD grant 2020.04453.BD to F.P. Further Arctic campaign logistic and traveling support was provided by the Portuguese Polar Program (PROPOLAR) and by the former Centre for Ice, Climate and Ecosystems at the Norwegian Polar Institute, the Research Council of Norway (project no. 244646), the Norwegian Ministries of Foreign Affairs and Climate and Environment through the program Arktis 2030 (project ID Arctic). This study was also partially funded by “Programa Operacional Regional de Lisboa” (Project N. 007317) and the Strategic Funding UIDB/04423/2020, UIDP/04565/2020, and UIDB/04565/2020 through national funds provided by FCT and European Regional Development Fund (ERDF), in the framework of the “PT2020” program.
Author information
Authors and Affiliations
Contributions
F.P., R.C., and C.M. conceptualized the manuscript. F.P. wrote the R code for all figures and tables and designed the Circos figures. F.P .wrote and R.C., C.M., P.A., and P.D. reviewed the first manuscript draft. C.M. and P.D. designed the sampling campaign. C.M., R.C., and P.A. funded the work. All authors reviewed, improved, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflicts of Interest
The authors declare no competing interests.
Supplementary Information
Supplementary Fig. S1
Quality scores profile for the forward 16S rRNA gene sequences (PNG 6307 kb)
Supplementary Fig. S2
Quality scores profile for the reverse 16S rRNA gene sequences (PNG 6497 kb)
Supplementary Fig. S3
MultiCoLA results of Procrustes and boxplot with the relative abundance equivalent of the selected cutoff, for non-rarefied data; a Procrustes results for the ASVs; b boxplots with relative abundance equivalent for each sample, after application of the selected threshold, for the ASVs; c Procrustes results for the OTUs; d boxplots with relative abundance equivalent for each sample, after application of the selected threshold, for the OTUs; e Procrustes results for the mOTUs; f boxplots with relative abundance equivalent for each sample, after application of the selected threshold, for the mOTUs (PNG 1425 kb)
Supplementary Fig. S4
MultiCoLA results of Procrustes for rarefied data. a Procrustes results for the ASVs; b Procrustes results for the OTUs; c Procrustes results for the mOTUs (PNG 323 kb)
Supplementary Fig. S5
a Rarefaction curve for ASV samples; b rarefaction curve for OTU samples; c rarefaction curves for mOTU samples (PNG 838 kb)
Supplementary Fig. S6
Alpha diversity for the total community (rarefied) and for the rare biosphere, defined by the MultiCoLA algorithm, across ASVs, OTUs, and mOTUs. a Number of sequences for the total community and b for the rare biosphere; c species richness for the total community and d rare biosphere; e Shannon equitability for the total community and f rare biosphere (PNG 583 kb)
Supplementary Fig. S7
Alpha diversity for the total community (non-rarefied) and for the rare biosphere, defined by the 0.1% relative abundance threshold, per sample, across ASVs, OTUs, and mOTUs. a Number of sequences for the total community and b for the rare biosphere; c species richness for the total community and d rare biosphere; e Shannon equitability for the total community and f rare biosphere (PNG 587 kb)
Supplementary Fig. S8
Alpha diversity for the total community (rarefied) and for the rare biosphere, defined by defined by the 0.1% relative abundance threshold, per sample, across ASVs, OTUs, and mOTUs. a Number of sequences for the total community and b for the rare biosphere; c species richness for the total community and d rare biosphere; e Shannon equitability for the total community and f rare biosphere (PNG 581 kb)
Supplementary Fig. S9
Dendrograms and multivariate analysis for the rare biosphere, defined by MultiCoLA and rarefied. a Dendrogram of the rare ASVs; b dendrogram of the rare OTUs; c dendrogram of the rare mOTUs; d CA for the rare ASVs; e CA for the rare OTUs; f CA for the rare mOTUs (PNG 1200 kb)
Supplementary Fig. S10
Dendrograms and multivariate analysis for the rare biosphere, defined by 0.1% relative abundance threshold, per sample, and non-rarefied. a Dendrogram of the rare ASVs; b dendrogram of the rare OTUs; c dendrogram of the rare mOTUs; d CA for the rare ASVs; e CA for the rare OTUs; f CA for the rare mOTUs (PNG 1187 kb)
Supplementary Fig. S11
Dendrograms and multivariate analysis for the rare biosphere, defined by 0.1% relative abundance threshold, per sample, and rarefied. a Dendrogram of the rare ASVs; b Dendrogram of the rare OTUs; c Dendrogram of the rare mOTUs; d CA for the rare ASVs; e CA for the rare OTUs; f CA for the rare mOTUs (PNG 1186 kb)
Supplementary Fig. S12
Stacked bar plots with the proportion of taxa for each type of rarity, if rare, and for the proportion of abundant taxa. Rare biosphere was defined by the MultiCoLA algorithm, from rarefied data. a The proportions for the rare ASVs; b the proportions for the rare OTUs; c the proportions for the rare mOTUs. The stacked bar plots are colored with purple for transiently rare taxa (TRT), blue for permanently rare taxa (PRT), green for conditionally rare taxa (CRT), and red for the abundant taxa. Within each dataset, different groups of samples were selected, specifically: “All” is for all samples; “March” is the group of samples from the month of March and they represent different depths (5, 50, and 250 m); “April” is the group of samples from the month of April and they represent different depths (5, 50, and 250 m); “June” is the group of samples from the month of June and they represent different depths (5, 20, and 250 m); “5m” is the group of samples taken at 5 m of depth and they represent the different months (March, April, and June); “20m or 50m” is the group of samples taken at 20 or 50 m of depth and they represent the different months (March, April, and June); “250m” is the group of samples taken at 250 m of depth and they represent the different months (March, April, and June) (PNG 260 kb)
Supplementary Fig. S13
Stacked bar plots with the proportion of taxa for each type of rarity, if rare, and for the proportion of abundant taxa. Rare biosphere was defined by the 0.1% relative abundance threshold, per sample, from non-rarefied data. a The proportions for the rare ASVs; b the proportions for the rare OTUs; c the proportions for the rare mOTUs. The stacked bar plots are colored with purple for transiently rare taxa (TRT), blue for permanently rare taxa (PRT), green for conditionally rare taxa (CRT), and red for the abundant taxa. Within each dataset, different groups of samples were selected, specifically: “All” is for all samples; “March” is the group of samples from the month of March and they represent different depths (5, 50, and 250 m); “April” is the group of samples from the month of April and they represent different depths (5, 50, and 250 m); “June” is the group of samples from the month of June and they represent different depths (5, 20, and 250 m); “5m” is the group of samples taken at 5 m of depth and they represent the different months (March, April, and June); “20m or 50m” is the group of samples taken at 20 or 50 m of depth and they represent the different months (March, April, and June); “250m” is the group of samples taken at 250 m of depth and they represent the different months (March, April, and June) (PNG 260 kb)
Supplementary Fig. S14
Stacked bar plots with the proportion of taxa for each type of rarity, if rare, and for the proportion of abundant taxa. Rare biosphere was defined by the 0.1% relative abundance threshold, per sample, from rarefied data. a The proportions for the rare ASVs; b the proportions for the rare OTUs; c the proportions for the rare mOTUs. The stacked bar plots are colored with purple for transiently rare taxa (TRT), blue for permanently rare taxa (PRT), green for conditionally rare taxa (CRT), and red for the abundant taxa. Within each dataset, different groups of samples were selected, specifically: “All” is for all samples; “March” is the group of samples from the month of March and they represent different depths (5, 50, and 250 m); “April” is the group of samples from the month of April and they represent different depths (5, 50, and 250 m); “June” is the group of samples from the month of June and they represent different depths (5, 20, and 250 m); “5m” is the group of samples taken at 5 m of depth and they represent the different months (March, April, and June); “20m or 50m” is the group of samples taken at 20 or 50 m of depth and they represent the different months (March, April, and June); “250m” is the group of samples taken at 250 m of depth and they represent the different months (March, April, and June) (PNG 263 kb)
Supplementary Data S1
Text file with the R code used for the processing of raw 16S rRNA gene sequences into ASVs and for the diversity metrics used for all datasets: ASVs, OTUs, and mOTUs (R 178 kb)
Supplementary Table S1
(XLSX 12.7 kb)
Rights and permissions
About this article
Cite this article
Pascoal, F., Costa, R., Assmy, P. et al. Exploration of the Types of Rarity in the Arctic Ocean from the Perspective of Multiple Methodologies. Microb Ecol 84, 59–72 (2022). https://doi.org/10.1007/s00248-021-01821-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01821-9