Abstract
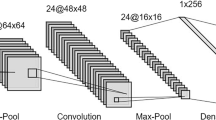
Determining the variety of wheat is important to know the physical and chemical properties which may be useful in grain processing. It also affects the price of wheat in the food industry. In this study, a Convolutional Neural Network (CNN)-based model was proposed to determine wheat varieties. Images of four different piles of wheat, two of which were the bread and the remaining durum wheat, were taken and image pre-processing techniques were applied. Small-sized images were cropped from high-resolution images, followed by data augmentation. Then, deep features were extracted from the obtained images using pre-trained seven different CNN models (AlexNet, ResNet18, ResNet50, ResNet101, Inceptionv3, DenseNet201, and Inceptionresnetv2). Support Vector Machines (SVM) classifier was used to classify deep features. The classification accuracies obtained by classification with various kernel functions such as Linear, Quadratic, Cubic and Gaussian were compared. The highest wheat classification accuracy was achieved with the deep features extracted with the Densenet201 model. In the classification made with the Cubic kernel function of SVM, the accuracy value was 98.1%.








Similar content being viewed by others
References
Dixon J, Braun H-J, Kosina P, Crouch JH (2009) Wheat facts and futures 2009. Cimmyt
Shewry PR, Hey SJ (2015) The contribution of wheat to human diet and health. Food Energy Security 4(3):178–202
FAO: http://www.fao.org/home/en/ (2019). Accessed
Lopes M, Reynolds M, Manes Y, Singh R, Crossa J, Braun H (2012) Genetic yield gains and changes in associated traits of CIMMYT spring bread wheat in a “historic” set representing 30 years of breeding. Crop Sci 52(3):1123–1131
Žilić S, Barać M, Pešić M, Dodig D, Ignjatović-Micić D (2011) Characterization of proteins from grain of different bread and durum wheat genotypes. Int J Mol Sci 12(9):5878–5894
Kimber G, Sears ER (1987) Evolution in the genus Triticum and the origin of cultivated wheat. Wheat and wheat improvement, vol 13. Soil Science Society of America, pp 154–164
Sayaslan A, Koyuncu M, Yildirim A, Güleç T, Sönmezoğlu ÖA, Kandemir N (2012) Some quality characteristics of selected durum wheat (Triticum durum) landraces. Turk J Agric For 36(6):749–756
Botwright T, Condon A, Rebetzke G, Richards R (2002) Field evaluation of early vigour for genetic improvement of grain yield in wheat. Aust J Agric Res 53(10):1137–1145
Dholakia B, Ammiraju J, Singh H, Lagu M, Röder M, Rao V et al (2003) Molecular marker analysis of kernel size and shape in bread wheat. Plant Breeding 122(5):392–395
Mahajan S, Das A, Sardana HK (2015) Image acquisition techniques for assessment of legume quality. Trends Food Sci Technology 42(2):116–133
Patrício DI, Rieder R (2018) Computer vision and artificial intelligence in precision agriculture for grain crops: a systematic review. Comput Electr Agric 153:69–81
Ferentinos KP (2018) Deep learning models for plant disease detection and diagnosis. Comput Electron Agric 145:311–318
Dong M, Mu S, Shi A, Mu W, Sun W (2020) Novel method for identifying wheat leaf disease images based on differential amplification convolutional neural network. Int J Agric Biol Eng 13(4):205–210
Jahan N, Flores P, Liu Z, Friskop A, Mathew JJ, Zhang Z (2020) Detecting and Distinguishing Wheat Diseases using Image Processing and Machine Learning Algorithms. 2020 ASABE Annual International Virtual Meeting. American Society of Agricultural and Biological Engineers, p 1
Manavalan R (2020) Automatic identification of diseases in grains crops through computational approaches: a review. Comput Electr Agric 178:105802
Basati Z, Jamshidi B, Rasekh M, Abbaspour-Gilandeh Y (2018) Detection of sunn pest-damaged wheat samples using visible/near-infrared spectroscopy based on pattern recognition. Spectrochimica Acta Part A 203:308–314
Davari A, Parker BL (2018) A review of research on Sunn Pest Eurygaster integriceps Puton (Hemiptera: Scutelleridae) management published 2004–2016. J Asia-Pacific Entomol 21(1):352–360
Huang L, Li T, Ding C, Zhao J, Zhang D, Yang G (2020) Diagnosis of the severity of fusarium head blight of wheat ears on the basis of image and spectral feature fusion. Sensors 20(10):2887
Sabanci K (2020) Detection of sunn pest-damaged wheat grains using artificial bee colony optimization-based artificial intelligence techniques. J Sci Food Agric 100(2):817–824
Hasan MM, Chopin JP, Laga H, Miklavcic SJ (2018) Detection and analysis of wheat spikes using convolutional neural networks. Plant Methods 14(1):1–13
Ma J, Li Y, Du K, Zheng F, Zhang L, Gong Z et al (2020) Segmenting ears of winter wheat at flowering stage using digital images and deep learning. Comput Electr Agric 168:105159
Rasti S, Bleakley CJ, Silvestre GC, Holden N, Langton D, O’Hare GM (2020) Crop growth stage estimation prior to canopy closure using deep learning algorithms. Neural Computing Applications, pp 1–11
Velumani K, Madec S, de Solan B, Lopez-Lozano R, Gillet J, Labrosse J et al (2020) An automatic method based on daily in situ images and deep learning to date wheat heading stage. Field Crop Res 252:107793
Belton D, Helmholz P, Long J, Zerihun A (2019) Crop height monitoring using a consumer-grade camera and UAV technology. PFG J Photogram Remote Sens Geoinfo Sci 87(5):249–262
Pound MP, Atkinson JA, Townsend AJ, Wilson MH, Griffiths M, Jackson AS et al (2017) Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. Gigascience. 6(10):gix083
Toda Y, Okura F, Ito J, Okada S, Kinoshita T, Tsuji H et al (2020) Training instance segmentation neural network with synthetic datasets for crop seed phenotyping. Commun Biol 3(1):1–12
Zhang C, Si Y, Lamkey J, Boydston RA, Garland-Campbell KA, Sankaran S (2018) High-throughput phenotyping of seed/seedling evaluation using digital image analysis. Agronomy 8(5):63
Adnan M, Abaid-ur-Rehman M, Latif MA, Ahmad N, Nazir M, Akhter N (2018) Mapping wheat crop phenology and the yield using machine learning (ML). Int J Adv Comput Sci Appl 9(8):301–306
Bijanzadeh E, Emam Y, Ebrahimie E (2010) Determining the most important features contributing to wheat grain yield using supervised feature selection model. Aust J Crop Sci 4(6):402–407
Kadir MKA, Ayob MZ, Miniappan N (2014) Wheat yield prediction: artificial neural network based approach. 2014 4th International Conference on Engineering Technology and Technopreneuship (ICE2T): IEEE, pp 161–165
Moghimi A, Yang C, Anderson JA (2020) Aerial hyperspectral imagery and deep neural networks for high-throughput yield phenotyping in wheat. Comput Electron Agric 172:105299
Olgun M, Onarcan AO, Özkan K, Işik Ş, Sezer O, Özgişi K et al (2016) Wheat grain classification by using dense SIFT features with SVM classifier. Comput Electron Agric 122:185–190
Sabanci K, Aslan MF, Durdu A (2020) Bread and durum wheat classification using wavelet based image fusion. J Sci Food Agric 100(15):5577–5585
Sabanci K, Kayabasi A, Toktas A (2017) Computer vision-based method for classification of wheat grains using artificial neural network. J Sci Food Agric 97(8):2588–2593
Taner A, Öztekin YB, Tekgüler A, Sauk H, Duran H (2018) Classification of varieties of grain species by artificial neural networks. Agronomy 8(7):123
Camargo A, Smith J (2009) An image-processing based algorithm to automatically identify plant disease visual symptoms. Biosys Eng 102(1):9–21
Sa I, Ge Z, Dayoub F, Upcroft B, Perez T, McCool C (2016) Deepfruits: a fruit detection system using deep neural networks. Sensors 16(8):1222
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7:1419
Özkan K, Işık Ş, Yavuz BT (2019) Identification of wheat kernels by fusion of RGB, SWIR, and VNIR samples. J Sci Food Agric 99(11):4977–4984
Zhang L, Sun H, Rao Z, Ji H (2020) Non-destructive identification of slightly sprouted wheat kernels using hyperspectral data on both sides of wheat kernels. Biosys Eng 200:188–199
Cinar I, Koklu M (2019) Classification of rice varieties using artificial intelligence methods. Int J Intell Syst Appl Eng 7(3):188–194
Bloice MD, Stocker C, Holzinger A (2017) Augmentor: an image augmentation library for machine learning. arXiv preprint arXiv:04680
Shorten C, Khoshgoftaar TM (2019) A survey on image data augmentation for deep learning. J Big Data 6(1):1–48
Lopes U, Valiati JF (2017) Pre-trained convolutional neural networks as feature extractors for tuberculosis detection. Comput Biol Med 89:135–143
Ismael AM, Şengür A (2021) Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst Appl 164:114054
Aslan MF, Unlersen MF, Sabanci K, Durdu A (2021) CNN-based transfer learning–BiLSTM network: a novel approach for COVID-19 infection detection. Appl Soft Comput 98:106912
Koklu M, Unlersen MF, Ozkan IA, Aslan MF, Sabanci K (2022) A CNN-SVM study based on selected deep features for grapevine leaves classification. Measurement 188:110425
Sabanci K, Aslan MF, Ropelewska E, Unlersen MF (2021) A convolutional neural network-based comparative study for pepper seed classification: analysis of selected deep features with support vector machine. J Food Process Eng e13955. https://doi.org/10.1111/jfpe.13955
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflict of interest.
Compliance with ethics requirements
This study does not contain any studies with human participants or animals performed by any authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Unlersen, M.F., Sonmez, M.E., Aslan, M.F. et al. CNN–SVM hybrid model for varietal classification of wheat based on bulk samples. Eur Food Res Technol 248, 2043–2052 (2022). https://doi.org/10.1007/s00217-022-04029-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-022-04029-4