Abstract
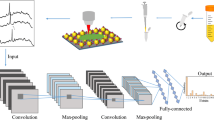
Carbapenem-resistant Enterobacteriaceae (CRE) is a major pathogen that poses a serious threat to human health. Unfortunately, currently, there are no effective measures to curb its rapid development. To address this, an in-depth study on the surface-enhanced Raman spectroscopy (SERS) of 22 strains of 7 categories of CRE using a gold silver composite SERS substrate was conducted. The residual networks with an attention mechanism to classify the SERS spectrum from three perspectives (pathogenic bacteria type, enzyme-producing subtype, and sensitive antibiotic type) were performed. The results show that the SERS spectrum measured by the composite SERS substrate was repeatable and consistent. The SERS spectrum of CRE showed varying degrees of species differences, and the strain difference in the SERS spectrum of CRE was closely related to the type of enzyme-producing subtype. The introduced attention mechanism improved the classification accuracy of the residual network (ResNet) model. The accuracy of CRE classification for different strains and enzyme-producing subtypes reached 94.0% and 96.13%, respectively. The accuracy of CRE classification by pathogen sensitive antibiotic combination reached 93.9%. This study is significant for guiding antibiotic use in CRE infection, as the sensitive antibiotic used in treatment can be predicted directly by measuring CRE spectra. Our study demonstrates the potential of combining SERS with deep learning algorithms to identify CRE without culture labels and classify its sensitive antibiotics. This approach provides a new idea for rapid and accurate clinical detection of CRE and has important significance for alleviating the rapid development of resistance to CRE.








Similar content being viewed by others
Data availability
The author confirms that all data generated or analyzed during the course of this study have been included in this article. The network model code involved in this article can be obtained through consultation with the corresponding author.
References
Potter RF, D’Souza AW, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Updat. 2016;29:30–46. https://doi.org/10.1016/j.drup.2016.09.002.
Armstrong T, Fenn SJ, Hardie KR. JMM Profile: Carbapenems: a broad-spectrum antibiotic. J Med Microbiol. 2021;70: 001462. https://doi.org/10.1099/jmm.0.001462.
Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543:15. https://doi.org/10.1038/nature.2017.21550.
Pokharel K, Dawadi BR, Bhatt CP, Gupte S, Jha B. Resistance pattern of carbapenem on Enterobacteriaceae. JNMA J Nepal Med Assoc. 2018;56:931–5. https://doi.org/10.31729/jnma.4006.
Morrill HJ, Pogue JM, Kaye KS, LaPlante KL. Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect Dis. 2015;2:ofv050. https://doi.org/10.1093/ofid/ofv050.
Lau AF, Wang H, Weingarten RA, Drake SK, Suffredini AF, Garfield MK, Chen Y, Gucek M, Youn J-H, Stock F, Tso H, DeLeo J, Cimino JJ, Frank KM, Dekker JP. A rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2014;52:2804–12. https://doi.org/10.1128/JCM.00694-14.
Moore NM, Cantón R, Carretto E, Peterson LR, Sautter RL, Traczewski MM, Carba-R Study Team. Rapid identification of five classes of carbapenem resistance genes directly from rectal swabs by use of the Xpert Carba-R assay. J Clin Microbiol. 2017;55:2268–75. https://doi.org/10.1128/JCM.00137-17.
Cheng C, Zheng F, Rui Y. Rapid detection of blaNDM, blaKPC, blaIMP, and blaVIM carbapenemase genes in bacteria by loop-mediated isothermal amplification. Microb Drug Resist. 2014;20:533–8. https://doi.org/10.1089/mdr.2014.0040.
Allen DM, Einarsson GG, Tunney MM, Bell SEJ. Characterization of bacteria using surface-enhanced Raman spectroscopy (SERS): influence of microbiological factors on the SERS spectra. Anal Chem. 2022;94:9327–35. https://doi.org/10.1021/acs.analchem.2c00817.
Qin Y-F, Lu X-Y, Shi Z, Huang Q-S, Wang X, Ren B, Cui L. Deep learning-enabled Raman spectroscopic identification of pathogen-derived extracellular vesicles and the biogenesis process. Anal Chem. 2022;94:12416–26. https://doi.org/10.1021/acs.analchem.2c02226.
Zhou X, Hu Z, Yang D, Xie S, Jiang Z, Niessner R, Haisch C, Zhou H, Sun P. Bacteria detection: from powerful SERS to its advanced compatible techniques. Adv Sci (Weinh). 2020;7:2001739. https://doi.org/10.1002/advs.202001739.
Wang L, Liu W, Tang J-W, Wang J-J, Liu Q-H, Wen P-B, Wang M-M, Pan Y-C, Gu B, Zhang X. Applications of Raman spectroscopy in bacterial infections: principles, advantages, and shortcomings. Front Microbiol. 2021;12: 683580. https://doi.org/10.3389/fmicb.2021.683580.
Li J, Wang C, Kang H, Shao L, Hu L, Xiao R, Wang S, Gu B. Label-free identification carbapenem-resistant Escherichia coli based on surface-enhanced resonance Raman scattering. RSC Adv. 2018;8:4761–5. https://doi.org/10.1039/C7RA13063E.
Liu W, Tang J-W, Lyu J-W, Wang J-J, Pan Y-C, Shi X-Y, Liu Q-H, Zhang X, Gu B, Wang L. Discrimination between carbapenem-resistant and carbapenem-sensitive Klebsiella pneumoniae strains through computational analysis of surface-enhanced Raman spectra: a pilot study. Microbiol Spectr. 2022;10:e02409-21. https://doi.org/10.1128/spectrum.02409-21.
Ho C-S, Jean N, Hogan CA, Blackmon L, Jeffrey SS, Holodniy M, Banaei N, Saleh AAE, Ermon S, Dionne J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat Commun. 2019;10:4927. https://doi.org/10.1038/s41467-019-12898-9.
Lu W, Chen X, Wang L, Li H, Fu YV. Combination of an artificial intelligence approach and laser tweezers Raman spectroscopy for microbial identification. Anal Chem. 2020;92:6288–96. https://doi.org/10.1021/acs.analchem.9b04946.
Lyu J-W, Zhang XD, Tang J-W, Zhao Y-H, Liu S-L, Zhao Y, Zhang N, Wang D, Ye L, Chen X-L, Wang L, Gu B. Rapid prediction of multidrug-resistant Klebsiella pneumoniae through deep learning analysis of SERS spectra. Microbiol Spectr. 2023;11:e0412622. https://doi.org/10.1128/spectrum.04126-22.
Zhao J, Lui H, McLean DI, Zeng H. Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy. Appl Spectrosc. 2007;61:1225–32. https://doi.org/10.1366/000370207782597003.
Guo M-H, Xu T-X, Liu J-J, Liu Z-N, Jiang P-T, Mu T-J, Zhang S-H, Martin RR, Cheng M-M, Hu S-M. Attention mechanisms in computer vision: a survey. Comp Visual Media. 2022;8:331–68. https://doi.org/10.1007/s41095-022-0271-y.
Choudhuri I, Khanra K, Maity P, Patra A, Maity GN, Pati BR, Nag A, Mondal S, Bhattacharyya N. Structure and biological properties of exopolysaccharide isolated from Citrobacter freundii. Int J Biol Macromol. 2021;168:537–49. https://doi.org/10.1016/j.ijbiomac.2020.12.063.
Mallakuntla MK, Vaikuntapu PR, Bhuvanachandra B, Das SN, Podile AR. Transglycosylation by a chitinase from Enterobacter cloacae subsp. cloacae generates longer chitin oligosaccharides. Sci Rep. 2017;7:5113. https://doi.org/10.1038/s41598-017-05140-3.
Dhinakaran AK, Dharmalingam P, Ganesh S, Venkatakrishnan K, Das S, Tan B. Molecular crosstalk between T cells and tumor uncovers GBM-specific T cell signatures in blood: noninvasive GBM diagnosis using immunosensors. ACS Nano. 2022;16:14134–48. https://doi.org/10.1021/acsnano.2c04160.
Zhang X, Wang X, Wang P, Fu Q, Zhu Z, Luo C, Chen J, Zhang Y, Li S. Facile synthesis of Ag-niobium ditelluride nanocomposites for the molecular fingerprint analysis of muscle tissues. J Mater Chem B. 2022;10:2944–51. https://doi.org/10.1039/d1tb02581c.
Sengupta A, Laucks ML, Davis EJ. Surface-enhanced Raman spectroscopy of bacteria and pollen. Appl Spectrosc. 2005;59:1016–23. https://doi.org/10.1366/0003702054615124.
Wang W, Kang S, Vikesland PJ. Surface-enhanced Raman spectroscopy of bacterial metabolites for bacterial growth monitoring and diagnosis of viral infection. Environ Sci Technol. 2021;55:9119–28. https://doi.org/10.1021/acs.est.1c02552.
Fu S, Wang X, Wang T, Li Z, Han D, Yu C, Yang C, Qu H, Chi H, Wang Y, Li S, Tian B, Li W, Xia Z. A sensitive and rapid bacterial antibiotic susceptibility test method by surface enhanced Raman spectroscopy. Braz J Microbiol. 2020;51:875–81. https://doi.org/10.1007/s42770-020-00282-5.
Cheng W-T, Liu M-T, Liu H-N, Lin S-Y. Micro-Raman spectroscopy used to identify and grade human skin pilomatrixoma. Microsc Res Tech. 2005;68:75–9. https://doi.org/10.1002/jemt.20229.
Zeiri L, Bronk BV, Shabtai Y, Eichler J, Efrima S. Surface-enhanced Raman spectroscopy as a tool for probing specific biochemical components in bacteria. Appl Spectrosc. 2004;58:33–40. https://doi.org/10.1366/000370204322729441.
Laska J, Widlarz J. Spectroscopic and structural characterization of low molecular weight fractions of polyaniline. Polymer. 2005;46:1485–95. https://doi.org/10.1016/j.polymer.2004.12.008.
Mircescu NE, Zhou H, Leopold N, Chiş V, Ivleva NP, Niessner R, Wieser A, Haisch C. Towards a receptor-free immobilization and SERS detection of urinary tract infections causative pathogens. Anal Bioanal Chem. 2014;406:3051–8. https://doi.org/10.1007/s00216-014-7761-4.
Rippa M, Castagna R, Sagnelli D, Vestri A, Borriello G, Fusco G, Zhou J, Petti L. SERS biosensor based on engineered 2D-aperiodic nanostructure for in-situ detection of viable Brucella bacterium in complex matrix. Nanomaterials (Basel). 2021;11:886. https://doi.org/10.3390/nano11040886.
Chan JW, Taylor DS, Zwerdling T, Lane SM, Ihara K, Huser T. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys J. 2006;90:648–56. https://doi.org/10.1529/biophysj.105.066761.
Dulyayangkul P, Wan Nur Ismah WAK, Douglas EJA, Avison MB. Mutation of kvrA causes OmpK35 and OmpK36 porin downregulation and reduced meropenem-vaborbactam susceptibility in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2020;64:e02208-e2219. https://doi.org/10.1128/AAC.02208-19.
Matsuoka T, Kawashima T, Nakamura T, Yabe T. Characterization and comparison of recombinant honeybee chymotrypsin-like protease (HCLPase) expressed in Escherichia coli and insect cells. Biosci Biotechnol Biochem. 2017;81:1401–4. https://doi.org/10.1080/09168451.2017.1318698.
Khan AU, Maryam L, Zarrilli R. Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 2017;17:101. https://doi.org/10.1186/s12866-017-1012-8.
Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends Microbiol. 2011;19:588–95. https://doi.org/10.1016/j.tim.2011.09.005.
Vera-Leiva A, Barría-Loaiza C, Carrasco-Anabalón S, Lima C, Aguayo-Reyes A, Domínguez M, Bello-Toledo H, González-Rocha G. KPC: Klebsiella pneumoniae carbapenemase, main carbapenemase in Enterobacteriaceae. Rev Chilena Infectol. 2017;34:476–84. https://doi.org/10.4067/S0716-10182017000500476.
Brandt C, Viehweger A, Singh A, Pletz MW, Wibberg D, Kalinowski J, Lerch S, Müller B, Makarewicz O. Assessing genetic diversity and similarity of 435 KPC-carrying plasmids. Sci Rep. 2019;9:11223. https://doi.org/10.1038/s41598-019-47758-5.
Lim FK, Liew YX, Cai Y, Lee W, Teo JQM, Lay WQ, Chung J, Kwa ALH. Treatment and outcomes of infections caused by diverse carbapenemase-producing carbapenem-resistant Enterobacterales. Front Cell Infect Microbiol. 2020;10: 579462. https://doi.org/10.3389/fcimb.2020.579462.
Abe R, Akeda Y, Sugawara Y, Matsumoto Y, Motooka D, Kawahara R, Yamamoto N, Tomono K, Iida T, Hamada S. Enhanced carbapenem resistance through multimerization of plasmids carrying carbapenemase genes. mBio. 2021;12:e0018621. https://doi.org/10.1128/mBio.00186-21.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 21876116, 82372353, and 81974299), the Guangdong Natural Science Foundation of China (No. 2021A1515011733 and 2023A1515010636), the GuangDong Basic and Applied Basic Research Foundation (2023A1515010938), the Guangdong Medical Science and Technology Research Found Project (B2023415), the Dongguan Science and Technology Commissioner Project (20231800500552), and the Social Development Science and Technology Project in Dongguan City (20211800900702).
Author information
Authors and Affiliations
Contributions
Shaoxin Li, Zhusheng Guo, and Yanjiao Zhang provided experimental design and technical assistance; Wen Wang and Ya Huang performed the experiments and analyzed data; Xin Wang and Xianglin Fang constructed the deep learning model; Wen Wang, Xin Wang, and Ya Huang wrote this paper; Luzhu Chen, Jingyi Zhong, and Ruoyi Li conducted preliminary research on the project; Ya Huang and Zhusheng Guo provided clinical bacterial samples; Yi Zhao provided technical assistance; Zhi Tang and Yanguang Cong were responsible for reviewing the manuscript. All authors discussed the results and contributed to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, W., Wang, X., Huang, Y. et al. Raman spectrum combined with deep learning for precise recognition of Carbapenem-resistant Enterobacteriaceae. Anal Bioanal Chem 416, 2465–2478 (2024). https://doi.org/10.1007/s00216-024-05209-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-024-05209-9