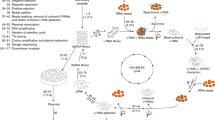
Abstract
A genome-inspired route to aptamer discovery that expands the sequence space beyond that available in traditional, combinatorial selection approaches is investigated for discovery of DNA-protein interactions in cancer. These interactions could then serve as the basis for new DNA aptamers to cancer-related proteins. The genome-inspired approach uses specific DNA sequences from the human genome to capture proteins from biological protein pools. The use of naturally occurring DNA sequences takes advantage of biological evolution of DNA sequences that bind to specific proteins to perform biological functions. Linking aptamer discovery to nature increa`ses the chances of uncovering protein-DNA affinity binding interactions that have biological significance as well as analytical utility. Here, the focus is on genomic, G-rich sequences that can form G-quadruplex (G4) structures. These structures are underrepresented in combinatorial libraries used for conventional aptamer selection. Additionally, G4-forming sequences are prone to inefficient PCR amplification, further biasing aptamer selection away from these structures. Nature provides a large diversity of G4-forming sequences throughout the human genome. They are prevalent in gene promoter regions, especially in oncogene promoters, and are therefore promising candidates for aptamers to regulatory proteins in cancer. The present work investigates protein capture from nuclear and cytoplasmic extracts of the breast cancer cell line MDA-MB-468 by G4-forming sequences from the CMYC, RB, and VEGF gene promoters. The studies included the effects of modifications of the VEGF sequence on the selectivity of protein capture, from which we identified promising aptamer candidates, subject to further refinement, to the proteins nucleolin and RPL19, both of which play important regulatory functions related to cancer.






Similar content being viewed by others
References
Adachi T, Nakamura Y. Aptamers: a review of their chemical properties and modifications for therapeutic application. Molecules. 2019;24(23):4229. https://doi.org/10.3390/molecules24234229.
Tombelli S, Minunni M, Mascini M. Analytical applications of aptamers. Biosens Bioelectron. 2005;20(12):2424–34. https://doi.org/10.1016/j.bios.2004.11.006.
McGown LB, Joseph MJ, Pitner JB, Vonk GP, Linn CP. The nucleic acid ligand. A new tool for molecular recognition. Anal Chem. 1995;67(21):663A–8A. https://doi.org/10.1021/ac00117a002.
Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–10. https://doi.org/10.1126/science.2200121.
Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–22. https://doi.org/10.1038/346818a0.
McManus SA, Li Y. Assessing the amount of quadruplex structures present within G2-tract synthetic random-sequence DNA libraries. PLoS One. 2013;8(5):e64131. https://doi.org/10.1371/journal.pone.0064131.
Gevertz J, Gan HH, Schlick T. In vitro RNA random pools are not structurally diverse: a computational analysis. RNA. 2005;11(6):853–63. https://doi.org/10.1261/rna.7271405.
Guschlbauer W, Chantot JF, Thiele D. Four-stranded nucleic acid structures 25 years later: from guanosine gels to telomer DNA. J Biomol Struct Dyn. 1990;8(3):491–511. https://doi.org/10.1080/07391102.1990.10507825.
Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34(19):5402–15. https://doi.org/10.1093/nar/gkl655.
Kwok CK, Merrick CJ. G-Quadruplexes: prediction, characterization, and biological application. Trends Biotechnol. 2017;35(10):997–1013. https://doi.org/10.1016/j.tibtech.2017.06.012.
Ric A, Ecochard V, Iacovoni JS, Boutonnet A, Ginot F, Ong-Meang V, et al. G-quadruplex aptamer selection using capillary electrophoresis-LED-induced fluorescence and Illumina sequencing. Anal Bioanal Chem. 2018;410(7):1991–2000. https://doi.org/10.1007/s00216-018-0865-5.
Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355(6360):564–6. https://doi.org/10.1038/355564a0.
Jellinek D, Green LS, Bell C, Janjić N. Inhibition of receptor binding by high-affinity RNA ligands to vascular endothelial growth factor. Biochemistry. 1994;33(34):10450–6. https://doi.org/10.1021/bi00200a028.
Lee JH, Canny MD, De Erkenez A, Krilleke D, Ng YS, Shima DT, et al. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc Natl Acad Sci U S A. 2005;102(52):18902–7. https://doi.org/10.1073/pnas.0509069102.
Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol. 2009;86(3):151–64. https://doi.org/10.1016/j.yexmp.2009.01.004.
Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35(2):406–13. https://doi.org/10.1093/nar/gkl1057 Erratum in: Nucleic Acids Res. 2007;35(6):2105.
Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43(18):8627–37. https://doi.org/10.1093/nar/gkv862.
Díaz-Fernández A, Miranda-Castro R. de-Los-Santos-Álvarez N, Lobo-Castañón MJ. Post-translational modifications in tumor biomarkers: the next challenge for aptamers? Anal Bioanal Chem. 2018;410(8):2059–65. https://doi.org/10.1007/s00216-018-0861-9.
Singer BS, Shtatland T, Brown D, Gold L. Libraries for genomic SELEX. Nucleic Acids Res. 1997;25(4):781–6. https://doi.org/10.1093/nar/25.4.781 Erratum in: Nucleic Acids Res 1997;25(21):4430.
Zimmermann B, Bilusic I, Lorenz C, Schroeder R. Genomic SELEX: a discovery tool for genomic aptamers. Methods. 2010;52(2):125–32. https://doi.org/10.1016/j.ymeth.2010.06.004.
Bevilacqua PC, Breen PC, Thaplyal P. Prospecting for aptamers in the human genome. Chem Biol. 2012;19(10):1218–20. https://doi.org/10.1016/j.chembiol.2012.10.001.
Zhang T, Zhang H, Wang Y, McGown LB. Capture and identification of proteins that bind to a GGA-rich sequence from the ERBB2 gene promoter region. Anal Bioanal Chem. 2012;404(6–7):1867–76. https://doi.org/10.1007/s00216-012-6322-y.
Albanese CM, Suttapitugsakul S, Perati S, McGown LB. A genome-inspired, reverse selection approach to aptamer discovery. Talanta. 2018;177:150–6. https://doi.org/10.1016/j.talanta.2017.08.093.
Connor AC, Frederick KA, Morgan EJ, McGown LB. Insulin capture by an insulin-linked polymorphic region G-quadruplex DNA oligonucleotide. J Am Chem Soc. 2006;128(15):4986–91. https://doi.org/10.1021/ja056097c.
Wang Y, Zhang H, Ligon LA, McGown LB. Association of insulin-like growth factor 2 with the insulin-linked polymorphic region in cultured fetal thymus cells. Biochemistry. 2009;48(34):8189–94. https://doi.org/10.1021/bi900958x.
Yoshida W, Mochizuki E, Takase M, Hasegawa H, Morita Y, Yamazaki H, et al. Selection of DNA aptamers against insulin and construction of an aptameric enzyme subunit for insulin sensing. Biosens Bioelectron. 2009;24(5):1116–20. https://doi.org/10.1016/j.bios.2008.06.016.
Xu Y, Sugiyama H. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb). Nucleic Acids Res. 2006;34(3):949–54. https://doi.org/10.1093/nar/gkj485.
Sun D, Guo K, Shin YJ. Evidence of the formation of G-quadruplex structures in the promoter region of the human vascular endothelial growth factor gene. Nucleic Acids Res. 2011;39(4):1256–65. https://doi.org/10.1093/nar/gkq926.
Yang D, Hurley LH. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides Nucleotides Nucleic Acids. 2006;25(8):951–68. https://doi.org/10.1080/15257770600809913.
González V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J Biol Chem. 2009;284(35):23622–35. https://doi.org/10.1074/jbc.M109.018028.
Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33(18):6070–80. https://doi.org/10.1093/nar/gki917.
Kaur H, Yung LY. Probing high affinity sequences of DNA aptamer against VEGF165. PLoS One. 2012;7(2):e31196. https://doi.org/10.1371/journal.pone.0031196.
Hasegawa H, Savory N, Abe K, Ikebukuro K. Methods for improving aptamer binding affinity. Molecules. 2016;21(4):421. https://doi.org/10.3390/molecules21040421.
Macdonald J, Houghton P, Xiang D, Duan W, Shigdar S. Truncation and mutation of a transferrin receptor aptamer enhances binding affinity. Nucleic Acid Ther. 2016;26(6):348–54. https://doi.org/10.1089/nat.2015.0585.
Gao S, Zheng X, Jiao B, Wang L. Post-SELEX optimization of aptamers. Anal Bioanal Chem. 2016;408(17):4567–73. https://doi.org/10.1007/s00216-016-9556-2.
Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34 (Web Server issue):W676–82. https://doi.org/10.1093/nar/gkl253.
Acknowledgements
We are grateful to Dr. Qishan Lin and Jinghua Zhu at the State University of New York at Albany Center for Functional Genomics for concentration, trypsin digestion, and LC-MS/MS analysis of our samples.
Funding
This work was supported by National Institutes of Health Grant 1R01GM112850-01A1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
N/A
Source of biological material
N/A
Statement on animal welfare
N/A
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 412 kb)
Rights and permissions
About this article
Cite this article
Morrissey, K.L., DeWitt, D., Shah, N. et al. Comparison of protein capture from a human cancer cell line by genomic G-quadruplex DNA sequences toward aptamer discovery. Anal Bioanal Chem 413, 3775–3788 (2021). https://doi.org/10.1007/s00216-021-03328-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03328-1