Abstract
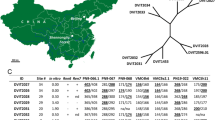
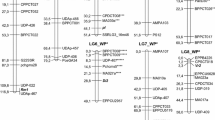
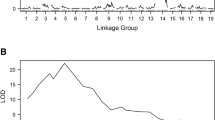
A population derived from a cross between grapevine breeding strain Gf.Ga-52-42 and cultivar ‘Solaris’ consisting of 265 F1-individuals was genetically mapped using SSR markers and screened for downy mildew resistance. Quantitative trait locus (QTL) analysis revealed two strong QTLs on linkage groups (LGs) 18 and 09. The locus on LG 18 was found to be identical with the previously described locus Rpv3 and is transmitted by Gf.Ga-52-42. ‘Solaris’ transmitted the resistance-related locus on LG 09 explaining up to 50% of the phenotypic variation in the population. This downy mildew resistance locus is named Rpv10 for resistance to Plasmopara viticola. Rpv10 was initially introgressed from Vitis amurensis, a wild species of the Asian Vitis gene pool. The one-LOD supported confidence interval of the QTL spans a section of 2.1 centi Morgan (cM) corresponding to 314 kb in the reference genome PN40024 (12x). Eight resistance gene analogues (RGAs) of the NBS–LRR type and additional resistance-linked genes are located in this region of PN40024. The F1 sub-population which contains the Rpv3 as well as the Rpv10 locus showed a significantly higher degree of resistance, indicating additive effects by pyramiding of resistance loci. Possibilities for using the resistance locus Rpv10 in a grapevine breeding programme are discussed. Furthermore, the marker data revealed ‘Severnyi’ × ‘Muscat Ottonel’ as the true parentage for the male parent of ‘Solaris’.






Similar content being viewed by others
References
Alleweldt G, Possingham JV (1988) Progress in grapevine breeding. Theor Appl Genet 75:669–673
Ampelografija of the USSR (1954) Volume III, Pishchepromizdat, Moscow, pp 23
Ampelografija of the USSR (1955) Volume V, Pishchepromizdat, Moscow, pp 259–265
Barker CL, Donald T, Pauquet J et al (2005) Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome library. Theor Appl Genet 111:370–377
Becker H (1981) Erste Ergebnisse der Züchtung interspezifischer Ertragssorten mit der Erbmasse der Vitis amurensis Ruprecht in Geisenheim. Deutsches Weinbau Jahrbuch:25–35
Becker N (2005) Zwei pilzresistente Weißweinsorten für den ökologischen Weinbau. http://www.landwirtschaft-bw.info. Accessed 11 January 2011
Bellin D, Peressott E, Merdinoglu D et al (2009) Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor Appl Genet 120:163–176
Blasi P, Schnee S, Wiedemann-Merdinoglu S et al. (2010) Genetic analysis of the resistance to downy and powdery mildews derived from cultivar Bronner. In: 6th international workshop of grapevine downy and powdery mildew
Blasi P, Blanc S, Wiedemann-Merdinoglu S et al. (2011) Construction of a reference linkage map of Vitis amurensis and genetic mapping of Rpv8, a locus conferring resistance to grapevine downy mildew. Theor Appl Genet pp 1–11
Boso S, Kassemeyer HH (2008) Different susceptibility of European grapevine cultivars for downy mildew. Vitis 47:39–49
Boso S, Martinez MC, Unger S, Kassemeyer HH (2006) Evaluation of foliar resistance to downy mildew in different cv. Albarino clones. Vitis 45:23–27
Bowers JE, Dangl GS, Vignani R, Meredith CP (1996) Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L). Genome 39:628–633
Bowers JE, Dangl GS, Meredith CP (1999) Development and characterization of additional microsatellite DNA markers for grape. Am J Enol Viticult 50:243–246
Bundessortenamt (2008) Beschreibende Sortenliste Reben. http://www.bundessortenamt.de
Burruano S (2000) The life-cycle of Plasmopara viticola, cause of downy mildew of vine. Mycologist 14:179–182
Cao H, Glazebrook J, Clarke JD et al (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88:57–63
Cipriani G, Marrazzo M, Di Gaspero G et al (2008) A set of microsatellite markers with long core repeat optimized for grape (Vitis spp.) genotyping. BMC Plant Biol 8:127
DeYoung BJ, Innes RW (2006) Plant NBS–LRR proteins in pathogen sensing and host defense. Nat Immunol 7:1243–1249
Di Gaspero G, Cipriani G (2003) Nucleotide binding site/leucine-rich repeats, Pto-like and receptor-like kinases related to disease resistance in grapevine. Mol Genet Genomics 269:612–623
Di Gaspero G, Cipriani G, Marrazzo MT et al (2005) Isolation of (AC)n-microsatellites in Vitis vinifera L. and analysis of genetic background in grapevines under marker assisted selection. Mol Breeding 15:11–20
Di Gaspero G, Cipriani G, Adam-Blondon AF, Testolin R (2007) Linkage maps of grapevine displaying the chromosomal locations of 420 microsatellite markers and 82 markers for R-gene candidates. Theor Appl Genet 114:1249–1263
Doligez A, Adam-Blondon AF, Cipriani G et al (2006) An integrated SSR map of grapevine based on five mapping populations. Theor Appl Genet 113:369–382
Donald TM, Pellerone F, Adam-Blondon AF et al (2002) Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor Appl Genet 104:610–618
Eibach R, Zyprian E, Welter L, Töpfer R (2007) The use of molecular markers for pyramiding resistance genes in grapevine breeding. Vitis 46:120–124
Eibach R, Hausmann L, Töpfer R (2010) Use of genetic diversity for grapevine resistance breeding. Mitt Klosterneuburg 60:332–337
Fischer BM, Salakhutdinov I, Akkurt M et al (2004) Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor Appl Genet 108:501–515
Gindro K, Pezet R, Viret O (2003) Histological study of the responses of two Vitis vinifera cultivars (resistant and susceptible) to Plasmopara viticola infections. Plant Physiol Bioch 41:846–853
Gindro K, Spring JL, Pezet R et al (2006) Histological and biochemical criteria for objective and early selection of grapevine cultivars resistant to Plasmopara viticola. Vitis 45:191–196
Hillebrand W, Lott H, Pfaff F (2003) Taschenbuch der Rebsorten. Fraund, Mainz
Hulbert SH, Ilott TW, Legg EJ et al (1988) Genetic analysis of the fungus Bremia lactucae, using restriction fragment length polymorphisms. Genetics 120:947–958
Hvarleva T, Bakalova A, Rusanov K et al (2009) Toward marker assisted selection for fungal disease resistance in grapevine. Biotechnol Biotec Eq 23:1431–1435
Ingle RA, Carstens M, Denby KJ (2006) PAMP recognition and the plant-pathogen arms race. Bioessays 28:880–889
Jaillon O, Aury JM, Noel B et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–468
Jansen RC (1993) Interval mapping of multiple quantitative trait loci. Genetics 135:205–211
Jürges G, Kassemeyer HH, Dürrenberger M et al (2009) The mode of interaction between Vitis and Plasmopara viticola Berk. & Curt. Ex de Bary depends on the host species. Plant Biol 11:886–898
Kiefer B, Riemann M, Buche C et al (2002) The host guides morphogenesis and stomatal targeting in the grapevine pathogen Plasmopara viticola. Planta 215:387–393
Kortekamp A, Wind R, Zyprian E (1998) Investigation of the interaction of Plasmopara viticola with susceptible and resistant grapevine cultivars. Z Pflanzenk Pflanzen 105:475–488
Kortekamp A, Welter L, Vogt S et al (2008) Identification, isolation and characterization of a CC–NBS–LRR candidate disease resistance gene family in grapevine. Mol Breeding 22:421–432
Le Henanff G, Heitz T, Mestre P et al (2009) Characterization of Vitis vinifera NPR1 homologs involved in the regulation of pathogenesis-related gene expression. BMC Plant Biol 9:54
Lott H, Pfaff F, Prior B (2010) Taschenbuch der Rebsorten. Fraund, Mainz, Germany
Luo S, He P, Zhou P, Zheng X (2001) Identification of molecular genetic markers tightly linked to downy mildew resistant genes in grape. Acta Genet Sinica 28:76–82
Marguerit E, Boury C, Manicki A et al (2009) Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine. Theor Appl Genet 118:1261–1278
Martins WS, Lucas DCS, Neves KF, Bertioli DJ (2009) WebSat—A web software for microsatellite marker development. Bioinformation 3:282–283
McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379
Merdinoglu D, Butterlin G, Bevilacqua L et al (2005) Development and characterization of a large set of microsatellite markers in grapevine (Vitis vinifera L.) suitable for multiplex PCR. Mol Breeding 15:349–366
Mohr HD (2005) Farbatlas Krankheiten. Schädlinge und Nützlinge an der Weinrebe. Ulmer, Stuttgart, Germany
Moreira F, Madini A, Marino R et al (2011) Genetic linkage maps of two interspecific grape crosses (Vitis spp.) used to localize quantitative trait loci for downy mildew resistance. Tree Genet Genomes 7:153–167
Nicholas P, Magarey P, Wachtel M (1994) Diseases and pests: grape production series no.1. Winetitels, Adelaide
OIV (2009) Descriptor list for grape varieties and Vitis species, 2nd edn. Office International de la Vigne et du Vin, Paris, http://www.oiv.org
Peressotti E, Wiedemann-Merdinoglu S, Delmotte F et al. (2010) Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol 10
Pezet R, Gindro K, Viret O, Spring JL (2004) Glycosylation and oxidative dimerization of resveratrol are respectively associated to sensitivity and resistance of grapevine cultivars to downy mildew. Physiol Mol Plant P 65:297–303
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365
Salmaso M, Malacarne G, Troggio M et al (2008) A grapevine (Vitis vinifera L.) genetic map integrating the position of 139 expressed genes. Theor Appl Genet 116:1129–1143
Scott KD, Eggler P, Seaton G et al (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Sefc KM, Regner F, Turetschek E et al (1999) Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 42:367–373
Staatliches Weinbauinstitut Freiburg (2010) Sorteninfoblatt Solaris. http://www.wbi-freiburg.de. Accessed 11 March 2011
Stein M, Dittgen J, Sanchez-Rodriguez C et al (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18:731–746
Thomas MR, Scott NS (1993) Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSs). Theor Appl Genet 86:985–990
Töpfer R, Hausmann L, Eibach R (2011) Molecular breeding. In: Adam-Blondon AF, Martinez-Zapater JM, Kole C (eds) Genetics, genomics and breeding of grapes. Science Publishers, Enfield, pp 160–185
Van Loon LC, Geraats BPJ, Linthorst HJM (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11:184–191
Van Ooijen JW (2004) MapQTL® 5, Software for the mapping of quantitative trait loci in experimental populations. Wageningen, Kyazma B.V
Van Ooijen JW (2006) JoinMap® 4, Software for calculation of genetic linkage maps in experimental populations. Wageningen, Kyazma B.V
Velasco R, Zharkikh A, Troggio M et al. (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. Plos One 2
Wan Y, Schwaninger H, He P, Wang Y (2007) Comparison of resistance to powdery mildew and downy mildew in Chinese wild grapes. Vitis 46:132–136
Wang XP, Wang YJ (2006) Isolation and diversity analysis of resistance gene analogs (RGAs) from wild Chinese Vitis species. Submitted May 2006 to the EMBL GenBank DDBJ databases
Welter LJ, Gokturk-Baydar N, Akkurt M et al (2007) Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol Breeding 20:359–374
Zhang JK, Hausmann L, Eibach R et al (2009) A framework map from grapevine V3125 (Vitis vinifera ‘Schiava grossa’ × Riesling’) × rootstock cultivar ‘Börner’ (Vitis riparia × Vitis cinerea) to localize genetic determinants of phylloxera root resistance. Theor Appl Genet 119:1039–1051
Acknowledgments
This project was funded by the “Forschungsring des Deutschen Weinbaus (FDW)”. We thank Erika Maul for the ampelographical support in the pedigree analysis and Friederike Rex for fruitful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Ordon.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2011_1695_MOESM2_ESM.emf
Suppl. Fig 1 a) Pedigree of `Solaris´ confirmed by the marker analysis. b) Currently assumed pedigree version (Staatliches Weinbauinstitut Freiburg 2010) and c) originally given pedigree version for `Solaris´ (Becker 1981). Continuous lines indicate SSR marker-confirmed linkages between tested cultivars (continuous boxes). Cultivars in dashed boxes and dashed lines are related predecessors according to the “Vitis International Variety Catalogue” (http://www.vivc.de). Dark grey represents presence of Rpv10, for `Saperavi Severnyi´ the presence or absence of Rpv10 is unknown. (EMF 365 kb)
122_2011_1695_MOESM3_ESM.emf
Suppl. Fig 2 Examples for SSR-Markers used for excluding `Zarya Severa´ as an ancestor of `Solaris´ (a, b) and confirming `Severnyi´ (a, b) and `Muscat Ottonel´ (c) as true ancestors. Observed fragment lengths (bp) are given for both alleles below the variety names. a) `Solaris´ inherited the allele with the 120 bp fragment from `Merzling´ which was confirmed as the maternal parent of `Solaris´ with all used markers. DNA of the pollen donor Geisenheim 6493 is not available because the name stands for a pollen mixture of a cross population. Thus, the 102 bp fragment of `Solaris´ must be found in one of the grandparents. In this case it is present in `Severnyi´. The absence in `Zarya Severa´ excludes this variety as an ancestor. b) Identical situation as demonstated in a) of a marker in the Rpv10 region. c) Example for a marker confirming `Muscat Ottonel´ as a grandparent of `Solaris´. `Saperavi severnyi´ can be excluded without testing its DNA by the observation that all alleles could be traced back to either `Severnyi´ or `Muscat Ottonel´. If `Saperavi severnyi´ was involved, up to ¼ (in case of only heterozygous markers) of the alleles (the ones going back to `Saperavi´) in `Solaris´ would not match. (EMF 181 kb)
Rights and permissions
About this article
Cite this article
Schwander, F., Eibach, R., Fechter, I. et al. Rpv10: a new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor Appl Genet 124, 163–176 (2012). https://doi.org/10.1007/s00122-011-1695-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1695-4