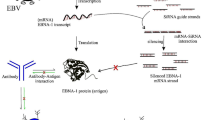
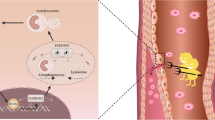
Abstract
Autoinflammation is the standard mechanism seen in systemic autoinflammatory disease (SAID) patients. This study aimed to investigate the effect of a candidate miRNA, miR-30e-3p, which was identified in our previous study, on the autoinflammation phenotype seen in SAID patients and to analyze its expression in a larger group of European SAID patients. We examined the potential anti-inflammatory effect of miR-30e-3p, which we had defined as one of the differentially expressed miRNAs in microarray analysis involved in inflammation-related pathways. This study validated our previous microarray results of miR-30e-3p in a cohort involving European SAID patients. We performed cell culture transfection assays for miR-30e-3p. Then, in transfected cells, we analyzed expression levels of pro-inflammatory genes; IL-1β, TNF-α, TGF-β, and MEFV. We also performed functional experiments, caspase-1 activation by fluorometric assay kit, apoptosis assay by flow cytometry, and cell migration assays by wound healing and filter system to understand the possible effect of miR-30e-3p on inflammation. Following these functional assays, 3'UTR luciferase activity assay and western blotting were carried out to identify the target gene of the aforementioned miRNA. MiR-30e-3p was decreased in severe European SAID patients like the Turkish patients. The functional assays associated with inflammation suggested that miR-30e-3p has an anti-inflammatory effect. 3'UTR luciferase activity assay demonstrated that miR-30e-3p directly binds to interleukin-1-beta (IL-1β), one of the critical molecules of inflammatory pathways, and reduces both RNA and protein levels of IL-1β. miR-30e-3p, which has been associated with IL-1β, a principal component of inflammation, might be of potential diagnostic and therapeutic value for SAIDs.
Key Messages
-
miR-30e-3p, which targets IL-1β, could have a role in the pathogenesis of SAID patients.
-
miR-30e-3p has a role in regulating inflammatory pathways like migration, caspase-1 activation.
-
miR-30e-3p has the potential to be used for future diagnostic and therapeutic approaches.






Similar content being viewed by others
Availability of data and materials
The graphical abstract was created with BioRender.com. The microarray data generated or analyzed during this study are included in this published article [14], but are available from the corresponding author on reasonable request.
References
Georgin-Lavialle S, Fayand A, Rodrigues F, Bachmeyer C, Savey L, Grateau G (2019) Autoinflammatory diseases: state of the art. Presse Med 48:e25–e48. https://doi.org/10.1016/j.lpm.2018.12.003
Harapas CR, Steiner A, Davidson S, Masters SL (2018) An update on autoinflammatory diseases: inflammasomopathies. Curr Rheumatol Rep 20:40. https://doi.org/10.1007/s11926-018-0750-4
McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, Mansfield E, Gadina M, Karenko L, Pettersson T et al (1999) Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell 97:133–144. https://doi.org/10.1016/s0092-8674(00)80721-7
Manthiram K, Zhou Q, Aksentijevich I, Kastner DL (2017) The monogenic autoinflammatory diseases define new pathways in human innate immunity and inflammation. Nat Immunol 18:832–842. https://doi.org/10.1038/ni.3777
van Kempen TS, Wenink MH, Leijten EF, Radstake TR, Boes M (2015) Perception of self: distinguishing autoimmunity from autoinflammation. Nat Rev Rheumatol 11:483–492. https://doi.org/10.1038/nrrheum.2015.60
Zhou X, Li X, Wu M (2018) miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct Target Ther 3:14. https://doi.org/10.1038/s41392-018-0006-9
Akkaya-Ulum YZ, Balci-Peynircioglu B, Karadag O, Eroglu FK, Kalyoncu U, Kiraz S, Ertenli AI, Özen S, Yilmaz E (2017) Alteration of the microRNA expression profile in familial Mediterranean fever patients. Clin Exp Rheumatol 35(Suppl 108):90–94
Akkaya-Ulum YZ, Akbaba TH, Tavukcuoglu Z, Chae JJ, Yilmaz E, Ozen S, Balci-Peynircioglu B (2021) Familial Mediterranean fever-related miR-197-3p targets IL1R1 gene and modulates inflammation in monocytes and synovial fibroblasts. Sci Rep 11:685. https://doi.org/10.1038/s41598-020-80097-4
Latsoudis H, Mashreghi MF, Grün JR, Chang HD, Stuhlmüller B, Repa A, Gergiannaki I, Kabouraki E, Vlachos GS, Häupl T et al (2017) Differential expression of miR-4520a associated with pyrin mutations in Familial Mediterranean Fever (FMF). J Cell Physiol 232:1326–1336. https://doi.org/10.1002/jcp.25602
Wada T, Toma T, Matsuda Y, Yachie A, Itami S, Taguchi YH, Murakami Y (2017) Microarray analysis of circulating microRNAs in familial Mediterranean fever. Mod Rheumatol 27:1040–1046. https://doi.org/10.1080/14397595.2017.1285845
Koga T, Migita K, Sato T, Sato S, Umeda M, Nonaka F, Fukui S, Kawashiri SY, Iwamoto N, Ichinose K et al (2018) MicroRNA-204-3p inhibits lipopolysaccharide-induced cytokines in familial Mediterranean fever via the phosphoinositide 3-kinase gamma pathway. Rheumatology (Oxford) 57:718–726. https://doi.org/10.1093/rheumatology/kex451
Lucherini OM, Obici L, Ferracin M, Fulci V, McDermott MF, Merlini G, Muscari I, Magnotti F, Dickie LJ, Galeazzi M et al (2013) First report of circulating microRNAs in tumour necrosis factor receptor-associated periodic syndrome (TRAPS). PLoS ONE 8:e73443–e73443. https://doi.org/10.1371/journal.pone.0073443
Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, Hornung V (2012) NLRP3 inflammasome activity is negatively controlled by miR-223. J Immunol 189:4175–4181. https://doi.org/10.4049/jimmunol.1201516
Akbaba TH, Akkaya-Ulum YZ, Tavukcuoglu Z, Bilginer Y, Ozen S, Balci-Peynircioglu B (2021) Inflammation-related differentially expressed common miRNAs in systemic autoinflammatory disorders patients can regulate the clinical course. Clin Exp Rheumatol
Gattorno M, Hofer M, Federici S, Vanoni F, Bovis F, Aksentijevich I, Anton J, Arostegui JI, Barron K, Ben-Cherit E et al (2019) Classification criteria for autoinflammatory recurrent fevers. Ann Rheum Dis 78:1025–1032. https://doi.org/10.1136/annrheumdis-2019-215048
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M (2021) KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 49:D545-d551. https://doi.org/10.1093/nar/gkaa970
Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD (2018) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47:D419–D426. https://doi.org/10.1093/nar/gky1038
Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM et al (2021) Gene set knowledge discovery with Enrichr. Curr Protocols 1:e90. https://doi.org/10.1002/cpz1.90
Consortium TGO (2020) The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 49:D325–D334. https://doi.org/10.1093/nar/gkaa1113
Clarke DJB, Jeon M, Stein DJ, Moiseyev N, Kropiwnicki E, Dai C, Xie Z, Wojciechowicz ML, Litz S, Hom J et al (2021) Appyters: turning Jupyter Notebooks into data-driven web apps. Patterns (N Y) 2:100213. https://doi.org/10.1016/j.patter.2021.100213
Otasek D, Morris JH, Bouças J, Pico AR, Demchak B (2019) Cytoscape automation: empowering workflow-based network analysis. Genome Biol 20:185. https://doi.org/10.1186/s13059-019-1758-4
Hoffman HM, Broderick L (2016) The role of the inflammasome in patients with autoinflammatory diseases. J Allergy Clin Immunol 138:3–14. https://doi.org/10.1016/j.jaci.2016.05.001
Awad F, Assrawi E, Louvrier C, Jumeau C, Georgin-Lavialle S, Grateau G, Amselem S, Giurgea I, Karabina SA (2018) Inflammasome biology, molecular pathology and therapeutic implications. Pharmacol Ther 187:133–149. https://doi.org/10.1016/j.pharmthera.2018.02.011
Lindsay MA (2008) microRNAs and the immune response. Trends Immunol 29:343–351. https://doi.org/10.1016/j.it.2008.04.004
O’Connell RM, Rao DS, Baltimore D (2012) microRNA regulation of inflammatory responses. Annu Rev Immunol 30:295–312. https://doi.org/10.1146/annurev-immunol-020711-075013
Luster AD, Alon R, von Andrian UH (2005) Immune cell migration in inflammation: present and future therapeutic targets. Nat Immunol 6:1182–1190. https://doi.org/10.1038/ni1275
Chen Y, Wang Z, Chen X, Peng X, Nie Q (2021) CircNFIC balances inflammation and apoptosis by sponging miR-30e-3p and regulating DENND1B expression. Genes (Basel) 12. https://doi.org/10.3390/genes12111829
Gramantieri L, Pollutri D, Gagliardi M, Giovannini C, Quarta S, Ferracin M, Casadei-Gardini A, Callegari E, De Carolis S, Marinelli S et al (2020) MiR-30e-3p influences tumor phenotype through MDM2/TP53 axis and predicts sorafenib resistance in hepatocellular carcinoma. Cancer Res 80:1720–1734. https://doi.org/10.1158/0008-5472.Can-19-0472
Gao K, Wang T, Qiao Y, Cui B (2021) MicroRNA-30e-3p inhibits glioma development and promotes drug sensitivity to temozolomide treatment via targeting canopy FGF signaling regulator 2. Cell Cycle 20:2361–2371. https://doi.org/10.1080/15384101.2021.1974789
Wang D, Zhu C, Zhang Y, Zheng Y, Ma F, Su L, Shao G (2017) MicroRNA-30e-3p inhibits cell invasion and migration in clear cell renal cell carcinoma by targeting Snail1. Oncol Lett 13:2053–2058. https://doi.org/10.3892/ol.2017.5690
Song A, Yang Y, He H, Sun J, Chang Q, Xue Q (2021) Inhibition of long non-coding RNA KCNQ1OT1 attenuates neuroinflammation and neuronal apoptosis through regulating NLRP3 expression via sponging miR-30e-3p. J Inflamm Res 14:1731–1742. https://doi.org/10.2147/jir.S291274
Funding
This work was supported by ERARE3 project (INSAID, grant number 003037603) and The Technical and Scientific Research Council of Turkey (TUBITAK), Grant number: 315S096. Three centers involved in this publication (IRCCS Istituto Giannina Gaslini of Genoa, University Hospital of Munster, and University Medical Center of Groninger) are members of the European Reference Network for Rare Immunodeficiency, Autoinflammatory and Autoimmune Diseases - Project ID No 739543.
Author information
Authors and Affiliations
Contributions
THA, ZYAU, SO, and BBP: conceptualization. THA and ZYAU: investigation. EDB, FP, HW, BK, MEVG, DF, MG, and SO: resources. THA and BBP: writing the original draft. THA, EDB, BBP, and SO: writing the review and editing. MEVG, DF, MG, SO, and BBP: funding acquisition. The authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The study was approved by the Hacettepe University Non-Interventional Clinical Research Ethics Board (GO 15/1744–19). Written consent was obtained from all parents and children.
Consent for publication
All authors agree on publishing.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Akbaba, T.H., Akkaya-Ulum, Y.Z., Batu, E.D. et al. Dysregulation of miRNA-30e-3p targeting IL-1β in an international cohort of systemic autoinflammatory disease patients. J Mol Med 101, 757–766 (2023). https://doi.org/10.1007/s00109-023-02327-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-023-02327-2