Abstract
Spinal muscular atrophy (SMA), a degenerative motor neuron disease and a leading cause of infant mortality, is caused by loss of functional survival motor neuron (SMN) protein due to SMN1 gene mutation. Here, using mouse and cell models for behavioral and histological studies, we found that SENP2 (SUMO/sentrin-specific protease 2)-deficient mice developed a notable SMA-like pathology phenotype with significantly decreased muscle fibers and motor neurons. At the molecular level, SENP2 deficiency in mice did not affect transcription but decreased SMN protein levels by promoting the SUMOylation of SMN. SMN was modified by SUMO2 with the E3 PIAS2α and deconjugated by SENP2. SUMOylation of SMN accelerated its degradation by the ubiquitin–proteasome degradation pathway with the ubiquitin E1 UBA1 (ubiquitin-like modifier activating enzyme 1) and E3 ITCH. SUMOylation of SMN increased its acetylation to inhibit the formation of Cajal bodies (CBs). These results showed that SENP2 deficiency induced hyper-SUMOylation of the SMN protein, which further affected the stability and functions of the SMN protein, eventually leading to the SMA-like phenotype. Thus, we uncovered the important roles for hyper-SUMOylation of SMN induced by SENP2 deficiency in motor neurons and provided a novel targeted therapeutic strategy for SMA.
Key messages
-
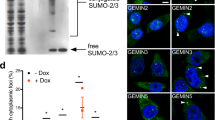
SENP2 deficiency enhanced the hyper-SUMOylation of SMN and promoted the degradation of SMN by the ubiquitin–proteasome pathway.
-
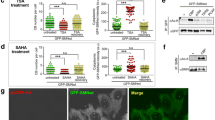
SUMOylation increased the acetylation of SMN to inhibit CB formation.
-
SENP2 deficiency caused hyper-SUMOylation of SMN protein, which further affected the stability and functions of SMN protein and eventually led to the occurrence of SMA-like pathology.








Similar content being viewed by others
Data availability
All the authors declare that data and materials are available upon request.
Code availability
Not applicable.
References
Rochette CF, Gilbert N, Simard LR (2001) SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum Genet 108:255–266. https://doi.org/10.1007/s004390100473
Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M et al (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–165. https://doi.org/10.1016/0092-8674(95)90460-3
Bergin A, Kim G, Price DL, Sisodia SS, Lee MK, Rabin BA (1997) Identification and characterization of a mouse homologue of the spinal muscular atrophy-determining gene, survival motor neuron. Gene 204:47–53. https://doi.org/10.1016/s0378-1119(97)00510-6
Cherry JJ, Androphy EJ (2012) Therapeutic strategies for the treatment of spinal muscular atrophy. Future Med Chem 4:1733–1750. https://doi.org/10.4155/fmc.12.107
Eggert C, Chari A, Laggerbauer B, Fischer U (2006) Spinal muscular atrophy: the RNP connection. Trends Mol Med 12:113–121. https://doi.org/10.1016/j.molmed.2006.01.005
Hamilton G, Gillingwater TH (2013) Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med 19:40–50. https://doi.org/10.1016/j.molmed.2012.11.002
Kim JK, Monani UR (2018) Augmenting the SMN protein to treat infantile spinal muscular atrophy. Neuron 97:1001–1003. https://doi.org/10.1016/j.neuron.2018.02.009
Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH (2009) Regulation of SMN protein stability. Mol Cell Biol 29:1107–1115. https://doi.org/10.1128/MCB.01262-08
Rademacher S, Detering NT, Schuning T, Lindner R, Santonicola P, Wefel IM, Dehus J, Walter LM, Brinkmann H, Niewienda A et al (2020) A single amino acid residue regulates PTEN-binding and stability of the spinal muscular atrophy protein SMN. Cells. https://doi.org/10.3390/cells9112405
Lunn MR, Wang CH (2008) Spinal muscular atrophy. Lancet 371:2120–2133. https://doi.org/10.1016/s0140-6736(08)60921-6
Tisdale S, Pellizzoni L (2015) Disease mechanisms and therapeutic approaches in spinal muscular atrophy. J Neurosci 35:8691–8700. https://doi.org/10.1523/JNEUROSCI.0417-15.2015
Wang CH, Finkel RS, Bertini ES, Schroth M, Simonds A, Wong B, Aloysius A, Morrison L, Main M, Crawford TO et al (2007) Consensus statement for standard of care in spinal muscular atrophy. J Child Neurol 22:1027–1049. https://doi.org/10.1177/0883073807305788
Hahnen E, Forkert R, Marke C, Rudnik-Schoneborn S, Schonling J, Zerres K, Wirth B (1995) Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum Mol Genet 4:1927–1933. https://doi.org/10.1093/hmg/4.10.1927
Monani UR, Sendtner M, Coovert DD, Parsons DW, Andreassi C, Le TT, Jablonka S, Schrank B, Rossoll W, Prior TW et al (2000) The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet 9:333–339. https://doi.org/10.1093/hmg/9.3.333
Lomeli H, Vazquez M (2011) Emerging roles of the SUMO pathway in development. Cell Mol Life Sci 68:4045–4064. https://doi.org/10.1007/s00018-011-0792-5
Hendriks IA, Vertegaal AC (2016) A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol 17:581–595. https://doi.org/10.1038/nrm.2016.81
Jackson SP, Durocher D (2013) Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 49:795–807. https://doi.org/10.1016/j.molcel.2013.01.017
Schwertman P, Bekker-Jensen S, Mailand N (2016) Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Biol 17:379–394. https://doi.org/10.1038/nrm.2016.58
Psakhye I, Castellucci F, Branzei D (2019) SUMO-chain-regulated proteasomal degradation timing exemplified in DNA replication initiation. Mol Cell 76(632–645):e636. https://doi.org/10.1016/j.molcel.2019.08.003
Yeh ET (2009) SUMOylation and De-SUMOylation: wrestling with life’s processes. J Biol Chem 284:8223–8227. https://doi.org/10.1074/jbc.R800050200
Nayak A, Muller S (2014) SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol 15:422. https://doi.org/10.1186/s13059-014-0422-2
Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, Cheng J, Yeh ET (2010) SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell 38:191–201. https://doi.org/10.1016/j.molcel.2010.03.005
Qi Y, Zuo Y, Yeh ET, Cheng J (2014) An essential role of small ubiquitin-like modifier (SUMO)-specific protease 2 in myostatin expression and myogenesis. J Biol Chem 289:3288–3293. https://doi.org/10.1074/jbc.M113.518282
Lee MH, Mabb AM, Gill GB, Yeh ET, Miyamoto S (2011) NF-kappaB induction of the SUMO protease SENP2: a negative feedback loop to attenuate cell survival response to genotoxic stress. Mol Cell 43:180–191. https://doi.org/10.1016/j.molcel.2011.06.017
Heo KS, Chang E, Le NT, Cushman H, Yeh ET, Fujiwara K, Abe J (2013) De-SUMOylation enzyme of sentrin/SUMO-specific protease 2 regulates disturbed flow-induced SUMOylation of ERK5 and p53 that leads to endothelial dysfunction and atherosclerosis. Circ Res 112:911–923. https://doi.org/10.1161/CIRCRESAHA.111.300179
Qi Y, Wang J, Bomben VC, Li DP, Chen SR, Sun H, Xi Y, Reed JG, Cheng J, Pan HL et al (2014) Hyper-SUMOylation of the Kv7 potassium channel diminishes the M-current leading to seizures and sudden death. Neuron 83:1159–1171. https://doi.org/10.1016/j.neuron.2014.07.042
Wu H, Chen X, Cheng J, Qi Y (2016) SUMOylation and potassium channels: links to epilepsy and sudden death. Adv Protein Chem Struct Biol 103:295–321. https://doi.org/10.1016/bs.apcsb.2015.11.009
Chen X, Zhang S, Huang J, Dong W, Xiao H, Shao H, Cheng J, Wu H, Qi Y (2018) Hyper-SUMOylation of K(+) channels in sudden unexplained death in epilepsy: isolation and primary culture of dissociated hippocampal neurons from newborn mice for subcellular localization. Methods Mol Biol 1684:63–71. https://doi.org/10.1007/978-1-4939-7362-0_6
Henley JM, Craig TJ, Wilkinson KA (2014) Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction. Physiol Rev 94:1249–1285. https://doi.org/10.1152/physrev.00008.2014
Guo C, Henley JM (2014) Wrestling with stress: roles of protein SUMOylation and deSUMOylation in cell stress response. IUBMB Life 66:71–77. https://doi.org/10.1002/iub.1244
Henley JM, Carmichael RE, Wilkinson KA (2018) Extranuclear SUMOylation in neurons. Trends Neurosci 41:198–210. https://doi.org/10.1016/j.tins.2018.02.004
Krumova P, Weishaupt JH (2013) Sumoylation in neurodegenerative diseases. Cell Mol Life Sci 70:2123–2138. https://doi.org/10.1007/s00018-012-1158-3
Anderson DB, Zanella CA, Henley JM, Cimarosti H (2017) Sumoylation: implications for neurodegenerative diseases. Adv Exp Med Biol 963:261–281. https://doi.org/10.1007/978-3-319-50044-7_16
Srivastav RK, Schwede S, Klaus M, Schwermann J, Gaestel M, Niedenthal R (2011) Monitoring protein-protein interactions in mammalian cells by trans-SUMOylation. Biochem J 438:495–503. https://doi.org/10.1042/BJ20110035
Nolle A, Zeug A, van Bergeijk J, Tonges L, Gerhard R, Brinkmann H, Al Rayes S, Hensel N, Schill Y, Apkhazava D et al (2011) The spinal muscular atrophy disease protein SMN is linked to the Rho-kinase pathway via profilin. Hum Mol Genet 20:4865–4878. https://doi.org/10.1093/hmg/ddr425
Tapia O, Lafarga V, Bengoechea R, Palanca A, Lafarga M, Berciano MT (2014) The SMN Tudor SIM-like domain is key to SmD1 and coilin interactions and to Cajal body biogenesis. J Cell Sci 127:939–946. https://doi.org/10.1242/jcs.138537
Yi Z, Wang S, Li L, Wu H, Ma Y, Qi Y, Pan H (2014) Diagnosis of MECP2 duplication syndrome with molecular genetic techniques. Zhonghua er ke za zhi = Chin J Pediatr 52:937–941
Palvimo JJ (2007) PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem Soc Trans 35:1405–1408. https://doi.org/10.1042/BST0351405
Rodriguez-Muela N, Parkhitko A, Grass T, Gibbs RM, Norabuena EM, Perrimon N, Singh R, Rubin LL (2018) Blocking p62-dependent SMN degradation ameliorates spinal muscular atrophy disease phenotypes. J Clin Invest 128:3008–3023. https://doi.org/10.1172/JCI95231
Geoffroy MC, Hay RT (2009) An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol 10:564–568. https://doi.org/10.1038/nrm2707
Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, Wikstrom M, Choudhary C, Houtsmuller AB, Marteijn JA et al (2013) RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 201:797–807. https://doi.org/10.1083/jcb.201212075
Erker Y, Neyret-Kahn H, Seeler JS, Dejean A, Atfi A, Levy L (2013) Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol Cell Biol 33:2163–2177. https://doi.org/10.1128/MCB.01019-12
Wishart TM, Mutsaers CA, Riessland M, Reimer MM, Hunter G, Hannam ML, Eaton SL, Fuller HR, Roche SL, Somers E et al (2014) Dysregulation of ubiquitin homeostasis and beta-catenin signaling promote spinal muscular atrophy. J Clin Invest 124:1821–1834. https://doi.org/10.1172/JCI71318
Han KJ, Foster D, Harhaj EW, Dzieciatkowska M, Hansen K, Liu CW (2016) Monoubiquitination of survival motor neuron regulates its cellular localization and Cajal body integrity. Hum Mol Genet 25:1392–1405. https://doi.org/10.1093/hmg/ddw021
Lafarga V, Tapia O, Sharma S, Bengoechea R, Stoecklin G, Lafarga M, Berciano MT (2018) CBP-mediated SMN acetylation modulates Cajal body biogenesis and the cytoplasmic targeting of SMN. Cell Mol Life Sci 75:527–546. https://doi.org/10.1007/s00018-017-2638-2
So BR, Wan L, Zhang Z, Li P, Babiash E, Duan J, Younis I, Dreyfuss G (2016) A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat Struct Mol Biol 23:225–230. https://doi.org/10.1038/nsmb.3167
Garvin AJ, Walker AK, Densham RM, Chauhan AS, Stone HR, Mackay HL, Jamshad M, Starowicz K, Daza-Martin M, Ronson GE et al (2019) The deSUMOylase SENP2 coordinates homologous recombination and nonhomologous end joining by independent mechanisms. Genes Dev 33:333–347. https://doi.org/10.1101/gad.321125.118
Liang Q, Zheng Q, Zuo Y, Chen Y, Ma J, Ni P, Cheng J (2019) SENP2 suppresses necdin expression to promote brown adipocyte differentiation. Cell Rep 28(2004–2011):e2004. https://doi.org/10.1016/j.celrep.2019.07.083
Qi Y, Yu Y, Li K, Chen Z, Liu Y, Zhang H (2014) Prevalence of obesity among primary students between 2009 to 2014 in China: a meta-analysis. Nutr Hosp 31:185–190. https://doi.org/10.3305/nh.2015.31.1.7967
Abera MB, Xiao J, Nofziger J, Titus S, Southall N, Zheng W, Moritz KE, Ferrer M, Cherry JJ, Androphy EJ et al (2016) ML372 blocks SMN ubiquitination and improves spinal muscular atrophy pathology in mice. JCI Insight 1:e88427. https://doi.org/10.1172/jci.insight.88427
Beltrao P, Bork P, Krogan NJ, van Noort V (2013) Evolution and functional cross-talk of protein post-translational modifications. Mol Syst Biol 9:714. https://doi.org/10.1002/msb.201304521
Funding
This research was funded by the National Natural Science Foundation of China (81671294 and 81870241 to YTQ) and the Fundamental Research Funds for the Central Universities (GK201903066 to HW).
Author information
Authors and Affiliations
Contributions
Y.H.Z. maintained the mice and identified the SMA-like phenotype; Y.H.Z., X.C., Q.Q.W., and Q.S. performed SUMOylation, ubiquitination, and acetylation analysis; Y.Y.Q., C.C.D., and D.Q.L. determined the SUMO sites of SMN; X.Y.Y., W.B.L., and X.L. performed the analysis of SMN1 transcript levels; Z.C.X., H.Y., and X.R. conducted the analysis of SMN protein levels; Y.J.S., Z.Z.Z., and Y.Z. conducted the Gemin2 experiments; H.M.W. and Y.T.Q. conceived the project; H.M.W. and Y.T.Q. wrote the paper with editorial input from Y.H.Z. and X.C.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Ethical approval
All animal care procedures and studies were performed according to protocols approved by the Institutional Animal Care and Use Committee of Shaanxi Normal University, Xi’an, China.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, Y., Chen, X., Wang, Q. et al. Hyper-SUMOylation of SMN induced by SENP2 deficiency decreases its stability and leads to spinal muscular atrophy-like pathology. J Mol Med 99, 1797–1813 (2021). https://doi.org/10.1007/s00109-021-02130-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-021-02130-x