Abstract
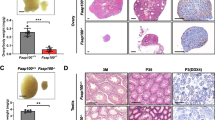
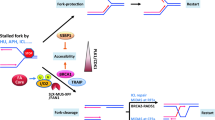
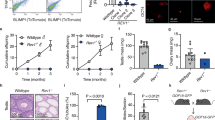
The proper development of primordial germ cells (PGCs) is an essential prerequisite for gametogenesis and mammalian fertility. The Fanconi anemia (FA) pathway functions in maintaining the development of PGCs. FANCT/UBE2T serves as an E2 ubiquitin-conjugating enzyme that ubiquitylates the FANCD2-FANCI complex to activate the FA pathway, but its role in the development of PGCs is not clear. In this study, we found that Ube2t knockout mice showed defects in PGC proliferation, leading to severe loss of germ cells after birth. Deletion of UBE2T exacerbated DNA damage and triggered the activation of the p53 pathway. We further demonstrated that UBE2T counteracted transcription-replication conflicts by resolving R-loops and stabilizing replication forks, and also protected common fragile sites by resolving R-loops in large genes and promoting mitotic DNA synthesis to maintain the genome stability of PGCs. Overall, these results provide new insights into the function and regulatory mechanisms of the FA pathway ensuring normal development of PGCs.










Similar content being viewed by others
Data availability
The sequencing data are available in Gene Expression Omnibus (GEO) database with the accession number GSE223410 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE223410). Enter token ujexukgqhtiltor into the box to review the data.
References
Findlay JK, Hutt KJ, Hickey M, Anderson RA (2015) How Is the number of primordial follicles in the ovarian reserve established. Biol Reprod 93:111
Bolcun-Filas E, Rinaldi VD, White ME, Schimenti JC (2014) Reversal of female infertility by Chk2 ablation reveals the oocyte DNA damage checkpoint pathway. Science 343:533–536
Musson R, Gąsior Ł, Bisogno S, Ptak GE (2022) DNA damage in preimplantation embryos and gametes: specification, clinical relevance and repair strategies. Hum Reprod Update 28:376–399
Ruth KS, Day FR, Hussain J et al (2021) Genetic insights into biological mechanisms governing human ovarian ageing. Nature 596:393–397
Tsui V, Crismani W (2019) The fanconi anemia pathway and fertility. Trends Genet 35:199–214
Luo Y, Hartford SA, Zeng R, Southard TL, Shima N, Schimenti JC (2014) Hypersensitivity of primordial germ cells to compromised replication-associated DNA repair involves ATM-p53-p21 signaling. PLoS Genet 10:e1004471
Hill RJ, Crossan GP (2019) DNA cross-link repair safeguards genomic stability during premeiotic germ cell development. Nat Genet 51:1283–1294
Ceccaldi R, Sarangi P, D’Andrea AD (2016) The fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol 17:337–349
Mamrak NE, Shimamura A, Howlett NG (2017) Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome fanconi anemia. Blood Rev 31:93–99
Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW (2005) The fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet 14:693–701
Luebben SW, Kawabata T, Johnson CS, O’Sullivan MG, Shima N (2014) A concomitant loss of dormant origins and FANCC exacerbates genome instability by impairing DNA replication fork progression. Nucleic Acids Res 42:5605–5615
Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M (2011) Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145:529–542
Schlacher K, Wu H, Jasin M (2012) A distinct replication fork protection pathway connects fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22:106–116
Gómez-González B, Aguilera A (2019) Transcription-mediated replication hindrance: a major driver of genome instability. Genes Dev 33:1008–1026
Roques C, Coulombe Y, Delannoy M et al (2009) MRE11-RAD50-NBS1 is a critical regulator of FANCD2 stability and function during DNA double-strand break repair. EMBO J 28:2400–2413
Yeo JE, Lee EH, Hendrickson EA, Sobeck A (2014) CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum Mol Genet 23:3695–3705
García-Muse T, Aguilera A (2016) Transcription-replication conflicts: how they occur and how they are resolved. Nat Rev Mol Cell Biol 17:553–563
García-Rubio ML, Pérez-Calero C, Barroso SI et al (2015) The fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet 11:e1005674
Schwab RA, Nieminuszczy J, Shah F et al (2015) The fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol Cell 60:351–361
Li S, Wu X (2020) Common fragile sites: protection and repair. Cell Biosci 10:29
Le TB, Millot GA, Blin ME, Brison O, Dutrillaux B, Debatisse M (2013) Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Rep 4:420–428
Debatisse M, Le TB, Letessier A, Dutrillaux B, Brison O (2012) Common fragile sites: mechanisms of instability revisited. Trends Genet 28:22–32
Zeman MK, Cimprich KA (2014) Causes and consequences of replication stress. Nat Cell Biol 16:2–9
El AE, Gerbault-Seureau M, Muleris M, Dutrillaux B, Debatisse M (2005) Premature condensation induces breaks at the interface of early and late replicating chromosome bands bearing common fragile sites. Proc Natl Acad Sci U S A 102:18069–18074
Minocherhomji S, Ying S, Bjerregaard VA et al (2015) Replication stress activates DNA repair synthesis in mitosis. Nature 528:286–290
Bhowmick R, Minocherhomji S, Hickson ID (2016) RAD52 facilitates mitotic DNA synthesis following replication stress. Mol Cell 64:1117–1126
Naim V, Wilhelm T, Debatisse M, Rosselli F (2013) ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat Cell Biol 15:1008–1015
Chan KL, Palmai-Pallag T, Ying S, Hickson ID (2009) Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11:753–760
Harrigan JA, Belotserkovskaya R, Coates J et al (2011) Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol 193:97–108
Lukas C, Savic V, Bekker-Jensen S et al (2011) 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13:243–253
Graber-Feesl CL, Pederson KD, Aney KJ, Shima N (2019) Mitotic DNA synthesis is differentially regulated between cancer and noncancerous cells. Mol Cancer Res 17:1687–1698
Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ (2007) UBE2T, the fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol 27:8421–8430
Lewis TW, Barthelemy JR, Virts EL et al (2019) Deficiency of the Fanconi anemia E2 ubiqitin conjugase UBE2T only partially abrogates Alu-mediated recombination in a new model of homology dependent recombination. Nucleic Acids Res 47:3503–3520
Spiller CM, Bowles J, Koopman P (2012) Regulation of germ cell meiosis in the fetal ovary. Int J Dev Biol 56:779–787
Vanni VS, Campo G, Cioffi R et al (2022) The neglected members of the family: non-BRCA mutations in the fanconi anemia/BRCA pathway and reproduction. Hum Reprod Update 28:296–311
Bremer S, Vogel R (1999) Pluripotent stem cells of the mouse as a potential in vitro model for mammalian germ cells. Sister chromatid exchanges induced by MMC and ENU in undifferentiated cell lines compared to differentiated cell lines. Mutat Res 444:97–102
Fischer M (2017) Census and evaluation of p53 target genes. Oncogene 36:3943–3956
Ceccaldi R, Parmar K, Mouly E et al (2012) Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell 11:36–49
Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M (2013) Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 32:340–353
Seki Y, Yamaji M, Yabuta Y et al (2007) Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134:2627–2638
García-Rubio M, Aguilera P, Lafuente-Barquero J et al (2018) Yra1-bound RNA-DNA hybrids cause orientation-independent transcription-replication collisions and telomere instability. Genes Dev 32:965–977
Aguilera A, García-Muse T (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46:115–124
Gan W, Guan Z, Liu J et al (2011) R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25:2041–2056
Naim V, Rosselli F (2009) The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol 11:761–768
Minocherhomji S, Hickson ID (2014) Structure-specific endonucleases: guardians of fragile site stability. Trends Cell Biol 24:321–327
Helmrich A, Ballarino M, Tora L (2011) Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell 44:966–977
Helmrich A, Stout-Weider K, Hermann K, Schrock E, Heiden T (2006) Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome Res 16:1222–1230
Krummel KA, Denison SR, Calhoun E, Phillips LA, Smith DI (2002) The common fragile site FRA16D and its associated gene WWOX are highly conserved in the mouse at Fra8E1. Genes Chromosomes Cancer 34:154–167
Helmrich A, Stout-Weider K, Matthaei A, Hermann K, Heiden T, Schrock E (2007) Identification of the human/mouse syntenic common fragile site FRA7K/Fra12C1–relation of FRA7K and other human common fragile sites on chromosome 7 to evolutionary breakpoints. Int J Cancer 120:48–54
Saitou M, Miyauchi H (2016) Gametogenesis from pluripotent stem cells. Cell Stem Cell 18:721–735
Tan H, Tee WW (2019) Committing the primordial germ cell: an updated molecular perspective. Wiley Interdiscip Rev Syst Biol Med 11:e1436
Holloway JK, Mohan S, Balmus G et al (2011) Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet 7:e1002094
Nadler JJ, Braun RE (2000) Fanconi anemia complementation group C is required for proliferation of murine primordial germ cells. Genesis 27:117–123
Wong JC, Alon N, Mckerlie C, Huang JR, Meyn MS, Buchwald M (2003) Targeted disruption of exons 1 to 6 of the fanconi anemia group a gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet 12:2063–2076
Hayashi Y, Otsuka K, Ebina M et al (2017) Distinct requirements for energy metabolism in mouse primordial germ cells and their reprogramming to embryonic germ cells. Proc Natl Acad Sci U S A 114:8289–8294
Leitch HG, Tang WW, Surani MA (2013) Primordial germ-cell development and epigenetic reprogramming in mammals. Curr Top Dev Biol 104:149–187
Nie Y, Wilson AF, DeFalco T, Meetei AR, Namekawa SH, Pang Q (2020) FANCD2 is required for the repression of germline transposable elements. Reproduction 159:659–668
Ying S, Minocherhomji S, Chan KL et al (2013) MUS81 promotes common fragile site expression. Nat Cell Biol 15:1001–1007
Wang K, Wang H, Li C et al (2021) Genomic profiling of native R loops with a DNA-RNA hybrid recognition sensor. Sci Adv. 7:eabe3516
Swain U, Subba RK (2011) Study of DNA damage via the comet assay and base excision repair activities in rat brain neurons and astrocytes during aging. Mech Ageing Dev 132:374–381
Bayona-Feliu A, Barroso S, Muñoz S, Aguilera A (2021) The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat Genet 53:1050–1063
Schwab RA, Niedzwiedz W (2011) Visualization of DNA replication in the vertebrate model system DT40 using the DNA fiber technique. J Vis Exp. https://doi.org/10.3791/3255
Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T (2014) deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42:W187-191
Acknowledgements
The authors thank Prof. Ping Zheng from Kunming Institute of Zoology, Chinese Academy of Sciences for providing technical support about DNA fiber assay and comet assay.
Funding
This work was supported by the National Key Research and Development Program of China [2022YFC2703800 and 2021YFC2700100]; Basic Science Center Program of NSFC [31988101]; National Natural Science Foundation for Distinguished Young Scholars [82125014]; National Natural Science Foundation of China [32170867 and 82071609]; Natural Science Foundation of Shandong Province for Grand Basic Projects [ZR2021ZD33]; Shandong Provincial Key Research and Development Program [2020ZLYS02]; Research Unit of Gametogenesis and Health of ART-Offspring; Chinese Academy of Medical Sciences [2020RU001]; Taishan Scholars Program for Young Experts of Shandong Province; and Qilu Young Scholars Program of Shandong University.
Author information
Authors and Affiliations
Contributions
Shidou Zhao, YQ, and JM designed the study. Y Yu performed most experiments; Y Yang completed the DNA fiber assay; WX completed the neutral comet assay; CW and Simin Zhao helped with the generation of MEFs; GL and RL participated in the genotyping; The manuscript was written by Y Yu and revised by Shidou Zhao, YQ and Z-JC.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
All animal experiments were conducted in accordance with the ethical guidelines approved by the Animal Care and Research Committee of Shandong University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, Y., Xu, W., Wen, C. et al. UBE2T resolves transcription-replication conflicts and protects common fragile sites in primordial germ cells. Cell. Mol. Life Sci. 80, 92 (2023). https://doi.org/10.1007/s00018-023-04733-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04733-8