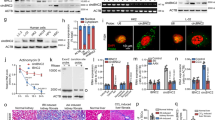
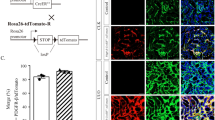
Abstract
Fibrosis is a relentlessly progressive and irreversible cause of organ damage, as in chronic kidney disease (CKD), but its underlying mechanisms remain elusive. We found that a circular RNA, circPTPN14, is highly expressed in human kidneys with biopsy-proved chronic interstitial fibrosis, mouse kidneys subjected to ischemia/reperfusion (IR) or unilateral ureteral obstruction (UUO), and TGFβ1-stimulated renal tubule epithelial cells (TECs). The intrarenal injection of circPTPN14 shRNA alleviated the progression of fibrosis in kidneys subjected to IR or UUO. Knockdown of circPTPN14 in TECs inhibited TGFβ1-induced expression of profibrotic genes, whereas overexpressing circPTPN14 increased the profibrotic effect of TGFβ1. The profibrotic action of circPTPN14 was ascribed to an increase in MYC transcription. The binding of circPTPN14 to the KH3 and KH4 domains of far upstream element (FUSE) binding protein 1 (FUBP1) enhanced the interaction between FUBP1 and FUSE domain, which was required for the initiation of MYC transcription. In human kidneys (n = 30) with biopsy-proved chronic interstitial fibrosis, the expression of circPTPN14 positively correlated with MYC expression. Taken together these studies show a novel mechanism in the pathogenesis of renal fibrosis, mediated by circPTPN14, which can be a target in the diagnosis and treatment of CKD.







Similar content being viewed by others
References
Rockey DC, Bell PD, Hill JA (2015) Fibrosis—A common pathway to organ injury and failure. N Engl J Med 372:1138–1149
Wynn TA, Ramalingam TR (2012) Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 18:1028–1040
Henderson NC, Rieder F, Wynn TA (2020) Fibrosis: from mechanisms to medicines. Nature 587:555–566
Boor P, Ostendorf T, Floege J (2010) Renal fibrosis: novel insights into mechanisms and therapeutic targets. Nat Rev Nephrol 6:643–656
Liu Y (2011) Cellular and molecular mechanisms of renal fibrosis. Nat Rev Nephrol 7:684–696
Chawla LS, Eggers PW, Star RA, Kimmel PL (2014) Acute kidney injury and chronic kidney disease as interconnected syndromes. N Engl J Med 371:58–66
Böttinger EP, Bitzer M (2002) TGF-ß signaling in renal disease. JASN 13:2600–2610
Varga J (1995) Modulation of collagen gene expression: its relation to fibrosis in systemic sclerosis and other disorders. Ann Intern Med 122:60
Kristensen LS et al (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:675–691
Chen L-L (2020) The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 21:475–490
Huang T et al (2020) Circular RNA YAP1 acts as the sponge of microRNA-21-5p to secure HK-2 cells from ischaemia/reperfusion-induced injury. J Cell Mol Med 24:4707–4715
Kölling M et al (2018) The Circular RNA ciRs-126 Predicts survival in critically ill patients with acute kidney injury. Kidney Int Rep 3:1144–1152
Xu H-P, Ma X-Y, Yang C (2021) Circular RNA TLK1 promotes sepsis-associated acute kidney injury by regulating inflammation and oxidative stress through miR-106a-5p/HMGB1 Axis. Front Mol Biosci 8:660269
Chen Z et al (2022) Circular RNA circPPP6R3 upregulates CD44 to promote the progression of clear cell renal cell carcinoma via sponging miR-1238-3p. Cell Death Dis 13:22
Mao W et al (2021) ciRS-7 is a prognostic biomarker and potential gene therapy target for renal cell carcinoma. Mol Cancer 20:142
Gong L-J, Wang X-Y, Yao X, Wu X, Gu W-Y (2021) CircESRP1 inhibits clear cell renal cell carcinoma progression through the CTCF-mediated positive feedback loop. Cell Death Dis 12:1081
Kölling M et al (2019) Circular RNAs in urine of kidney transplant patients with acute T Cell-mediated allograft rejection. Clin Chem 65:1287–1294
Wang J et al (2021) MicroRNA-874-3p/ADAM (a disintegrin and metalloprotease) 19 mediates macrophage activation and renal fibrosis after acute kidney injury. Hypertension 77:1613–1626
Wang P et al (2018) Long noncoding RNA lnc-TSI inhibits renal fibrogenesis by negatively regulating the TGF-β/Smad3 pathway. Sci Transl Med 10:2039
Wang J et al (2022) Inhibition of METTL3 attenuates renal injury and inflammation by alleviating TAB3 m6A modifications via IGF2BP2-dependent mechanisms. Sci Transl Med 14:2709
Jiang L et al (2022) METTL3-mediated m6A modification of TIMP2 mRNA promotes podocyte injury in diabetic nephropathy. Mol Ther 30:1721–1740
Thomson DW, Dinger ME (2016) Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17:272–283
Hansen TB et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388
Shi L et al (2021) A tumor-suppressive circular RNA mediates uncanonical integrin degradation by the proteasome in liver cancer. Sci Adv 7:5043
Chen Z et al (2021) Circular RNA cia-MAF drives self-renewal and metastasis of liver tumor-initiating cells via transcription factor MAFF. J Clin Investig 131:e148020
Shen S et al (2021) circPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann Rheum Dis 80:1209–1219
Duncan R et al (1994) A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev 8:465–480
Liu J et al (2006) The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J 25:2119–2130
Edeling M, Ragi G, Huang S, Pavenstädt H, Susztak K (2016) Developmental signalling pathways in renal fibrosis: the roles of Notch Wnt and Hedgehog. Nat Rev Nephrol 12:426–439
Jeremy Wen Q et al (2015) Targeting megakaryocytic-induced fibrosis in myeloproliferative neoplasms by AURKA inhibition. Nat Med 21:1473–1480
Shen Y et al (2017) c-Myc promotes renal fibrosis by inducing integrin αv-mediated transforming growth factor-β signaling. Kidney Int 92:888–899
Pelengaris S, Khan M, Evan G (2002) c-MYC: more than just a matter of life and death. Nat Rev Cancer 2:764–776
Wang H et al (2007) Improved low molecular weight Myc-Max inhibitors. Mol Cancer Ther 6:2399–2408
Duncan R, Collins I, Tomonaga T, Zhang T, Levens D (1996) A unique transactivation sequence motif is found in the carboxyl-terminal domain of the single-strand-binding protein FBP. Mol Cell Biol 16:2274–2282
Braddock DT (2002) Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J 21:3476–3485
van Zonneveld AJ, Kölling M, Bijkerk R, Lorenzen JM (2021) Circular RNAs in kidney disease and cancer. Nat Rev Nephrol 17:814–826
Kristensen LS, Jakobsen T, Hager H, Kjems J (2022) The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol 19:188–206
Davis AC, Wims M, Spotts GD, Hann SR, Bradley A (1993) A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev 7:671–682
Zhou W et al (2016) Far Upstream element binding protein plays a crucial role in embryonic development hematopoiesis and stabilizing myc expression levels. The Am J Pathol 186(3) 701-715. https://doi.org/10.1016/j.ajpath.2015.10.028
Kress TR, Sabò A, Amati B (2015) MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer 15:593–607
Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV (2015) MYC, metabolism, and cancer. Cancer Discov 5:1024–1039
Valiente-Alandi I et al (2018) Inhibiting fibronectin attenuates fibrosis and improves cardiac function in a model of heart failure. Circulation 138:1236–1252
Winkler M et al (2021) Endothelial GATA4 controls liver fibrosis and regeneration by preventing a pathogenic switch in angiocrine signaling. J Hepatol 74:380–393
Nevzorova YA et al (2013) Overexpression of c-myc in hepatocytes promotes activation of hepatic stellate cells and facilitates the onset of liver fibrosis. Biochim Biophys Acta Mol Basis Dis 1832:1765–1775
Kang HM et al (2015) Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med 21:37–46
Hofacker IL, Stadler PF (2006) Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics 22:1172–1176
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Popenda M et al (2012) Automated 3D structure composition for large RNAs. Nucleic Acids Res 40:e112–e112
Yan Y, Zhang D, Zhou P, Li B, Huang S-Y (2017) HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 45:W365–W373
Acknowledgements
We thank Prof. Pedro A. Jose and Prof. Ines Armando (The George Washington University, USA) for precious suggestions and draft revising. We are grateful for the technical support by the Core Facilities, Zhejiang University School of Medicine, and the Center of Cryo-Electron Microscopy of Zhejiang University.
Funding
This study was supported by the funding from Primary Research and Development Plan of Zhejiang Province (2020C03034) to Fei Han, and National Natural Science Foundation of China (81770674) to Fei Han.
Author information
Authors and Affiliations
Contributions
F.H., W.L., W.N., M.L. and B.L. conceived and designed the research. W.N., M.L. and B.L. performed most of the experiments, with the assistance from Y.Z., Y.W., J.W., L.J. and A.N., under the supervision of F.H.. L.X., X.Z.S., J.C. provided administrative support and suggestion. W.N., M.L. and B.L. wrote the original manuscript, with reviewing and editing by F.H.. All authors read and approved this version of manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nie, W., Li, M., Liu, B. et al. A circular RNA, circPTPN14, increases MYC transcription by interacting with FUBP1 and exacerbates renal fibrosis. Cell. Mol. Life Sci. 79, 595 (2022). https://doi.org/10.1007/s00018-022-04603-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04603-9