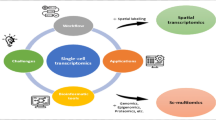
Abstract
Single-cell sequencing is widely used in biological and medical studies. However, its application with multiple samples is hindered by inefficient sample processing, high experimental costs, ambiguous identification of true single cells, and technical batch effects. Here, we introduce sample-multiplexing approaches for single-cell sequencing in transcriptomics, epigenomics, genomics, and multiomics. In single-cell transcriptomics, sample multiplexing uses variants of native or artificial features as sample markers, enabling sample pooling and decoding. Such features include: (1) natural genetic variation, (2) nucleotide-barcode anchoring on cellular or nuclear membranes, (3) nucleotide-barcode internalization to the cytoplasm or nucleus, (4) vector-based barcode expression in cells, and (5) nucleotide-barcode incorporation during library construction. Other single-cell omics methods are based on similar concepts, particularly single-cell combinatorial indexing. These methods overcome current challenges, while enabling super-loading of single cells. Finally, selection guidelines are presented that can accelerate technological application.





Similar content being viewed by others
Availability of data and material
Not applicable.
References
Tang F et al (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6:377–382. https://doi.org/10.1038/nmeth.1315
Shalek AK et al (2013) Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498:236–240. https://doi.org/10.1038/nature12172
Tang F et al (2011) Development and applications of single-cell transcriptome analysis. Nat Methods 8:S6-11. https://doi.org/10.1038/nmeth.1557
Deng Q et al (2014) Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science (New York, N.Y.) 343:193–196. https://doi.org/10.1126/science.1245316
Zhang X et al (2016) Single-cell sequencing for precise cancer research: progress and prospects. Can Res 76:1305–1312. https://doi.org/10.1158/0008-5472.Can-15-1907
Suvà ML, Tirosh I (2019) Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell 75:7–12. https://doi.org/10.1016/j.molcel.2019.05.003
Wen L, Tang F (2016) Single-cell sequencing in stem cell biology. Genome Biol 17:71. https://doi.org/10.1186/s13059-016-0941-0
Chen H et al (2019) Revolutionizing immunology with single-cell RNA sequencing. Cell Mol Immunol 16:242–249. https://doi.org/10.1038/s41423-019-0214-4
Ofengeim D et al (2017) Single-cell RNA sequencing: unraveling the brain one cell at a time. Trends Mol Med 23:563–576. https://doi.org/10.1016/j.molmed.2017.04.006
Cuevas-Diaz Duran R et al (2017) Single-cell RNA-sequencing of the brain. Clin Transl Med 6:20. https://doi.org/10.1186/s40169-017-0150-9
Mu Q et al (2019) Deciphering brain complexity using single-cell sequencing. Genomics Proteomics Bioinformatics 17:344–366. https://doi.org/10.1016/j.gpb.2018.07.007
Tšuiko O et al (2020) Preimplantation Genetic Testing: single-cell technologies at the forefront of PGT and embryo research. Reproduction (Cambridge, England) 160:A19-a31. https://doi.org/10.1530/rep-20-0102
Shangguan Y et al (2020) Application of single-cell RNA sequencing in embryonic development. Genomics 112:4547–4551. https://doi.org/10.1016/j.ygeno.2020.08.007
Peng G et al (2020) Using single-cell and spatial transcriptomes to understand stem cell lineage specification during early embryo development. Annu Rev Genomics Hum Genet 21:163–181. https://doi.org/10.1146/annurev-genom-120219-083220
Hedlund E, Deng Q (2018) Single-cell RNA sequencing: technical advancements and biological applications. Mol Aspects Med 59:36–46. https://doi.org/10.1016/j.mam.2017.07.003
Huang X et al (2018) High throughput single cell RNA sequencing, bioinformatics analysis and applications. Adv Exp Med Biol 1068:33–43. https://doi.org/10.1007/978-981-13-0502-3_4
Yasen A et al (2020) Progress and applications of single-cell sequencing techniques. Infect Genet Evol 80:104198. https://doi.org/10.1016/j.meegid.2020.104198
Zheng GX et al (2017) Massively parallel digital transcriptional profiling of single cells. Nat Commun 8:14049. https://doi.org/10.1038/ncomms14049
Shum EY et al (2019) Quantitation of mRNA transcripts and proteins using the BD Rhapsody™ single-cell analysis system. Adv Exp Med Biol 1129:63–79. https://doi.org/10.1007/978-981-13-6037-4_5
Svensson V et al (2018) Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc 13(4):599–604. https://doi.org/10.1038/nprot.2017.149
Valihrach L et al (2018) Platforms for single-cell collection and analysis. Int J Mol Sci. https://doi.org/10.3390/ijms19030807
Regev A et al (2017) The human cell Atlas. Elife. https://doi.org/10.7554/eLife.27041
Li H et al (2021) Fly Cell Atlas: a single-cell transcriptomic atlas of the adult fruit fly. J bioRxiv. https://doi.org/10.1101/2021.07.04.451050
Rhee SY et al (2019) Towards building a plant cell Atlas. Trends Plant Sci 24:303–310. https://doi.org/10.1016/j.tplants.2019.01.006
Wolock SL et al (2019) Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst 8(4):281–291. https://doi.org/10.1016/j.cels.2018.11.005 (e9)
McGinnis CS et al (2019) DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 8(4):329-337.e4. https://doi.org/10.1016/j.cels.2019.03.003
Hicks SC et al (2018) Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics (Oxford, England) 19:562–578. https://doi.org/10.1093/biostatistics/kxx053
Kang HM et al (2018) Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 36:89–94. https://doi.org/10.1038/nbt.4042
Xu J et al (2019) Genotype-free demultiplexing of pooled single-cell RNA-seq. Genome Biol 20:290. https://doi.org/10.1186/s13059-019-1852-7
Huang Y et al (2019) Vireo: Bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol 20:273. https://doi.org/10.1186/s13059-019-1865-2
Heaton H et al (2020) Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat Methods 17:615–620. https://doi.org/10.1038/s41592-020-0820-1
Stoeckius M et al (2017) Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14:865–868. https://doi.org/10.1038/nmeth.4380
Peterson VM et al (2017) Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol 35:936–939. https://doi.org/10.1038/nbt.3973
Stoeckius M et al (2018) Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 19:224. https://doi.org/10.1186/s13059-018-1603-1
Wu H et al (2019) Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol 30:23–32. https://doi.org/10.1681/asn.2018090912
Slyper M et al (2020) A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med 26:792–802. https://doi.org/10.1038/s41591-020-0844-1
Ding J et al (2020) Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 38:737–746. https://doi.org/10.1038/s41587-020-0465-8
Habib N et al (2017) Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14:955–958. https://doi.org/10.1038/nmeth.4407
Nagy C et al (2018) Single-nucleus RNA sequencing shows convergent evidence from different cell types for altered synaptic plasticity in major depressive disorder
Lake BB et al (2018) Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol 36:70–80. https://doi.org/10.1038/nbt.4038
Gaublomme JT et al (2019) Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat Commun 10:2907. https://doi.org/10.1038/s41467-019-10756-2
Weber RJ et al (2014) Efficient targeting of fatty-acid modified oligonucleotides to live cell membranes through stepwise assembly. Biomacromol 15:4621–4626. https://doi.org/10.1021/bm501467h
McGinnis CS et al (2019) MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods 16:619–626. https://doi.org/10.1038/s41592-019-0433-8
Fang L et al (2021) CASB: a concanavalin A-based sample barcoding strategy for single-cell sequencing. Mol Syst Biol 17:e10060. https://doi.org/10.15252/msb.202010060
Gehring J et al (2020) Highly multiplexed single-cell RNA-seq by DNA oligonucleotide tagging of cellular proteins. Nat Biotechnol 38:35–38. https://doi.org/10.1038/s41587-019-0372-z
Shin D et al (2019) Multiplexed single-cell RNA-seq via transient barcoding for simultaneous expression profiling of various drug perturbations. Sci Adv 5:eaav2249. https://doi.org/10.1126/sciadv.aav2249
Srivatsan SR et al (2020) Massively multiplex chemical transcriptomics at single-cell resolution. Science (New York, N.Y.) 367:45–51. https://doi.org/10.1126/science.aax6234
Guo C et al (2019) Cell Tag Indexing: genetic barcode-based sample multiplexing for single-cell genomics. Genome Biol 20:90. https://doi.org/10.1186/s13059-019-1699-y
Dixit A et al (2016) Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167:1853-1866.e1817. https://doi.org/10.1016/j.cell.2016.11.038
Adamson B et al (2016) A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167:1867–1882. https://doi.org/10.1016/j.cell.2016.11.048 (e1821)
Jaitin DA et al (2016) Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-Seq. Cell 167:1883–1896. https://doi.org/10.1016/j.cell.2016.11.039 (e1815)
Datlinger P et al (2017) Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods 14:297–301. https://doi.org/10.1038/nmeth.4177
Wagner DE, Klein AM (2017) Genetic screening enters the single-cell era. Nat Methods 14:237–238. https://doi.org/10.1038/nmeth.4196
Aarts M et al (2017) Coupling shRNA screens with single-cell RNA-seq identifies a dual role for mTOR in reprogramming-induced senescence. Genes Dev 31:2085–2098. https://doi.org/10.1101/gad.297796.117
Uzbas F et al (2019) BART-Seq: cost-effective massively parallelized targeted sequencing for genomics, transcriptomics, and single-cell analysis. Genome Biol 20:155. https://doi.org/10.1186/s13059-019-1748-6
Datlinger P et al (2021) Ultra-high-throughput single-cell RNA sequencing and perturbation screening with combinatorial fluidic indexing. Nat Methods 18:635–642. https://doi.org/10.1038/s41592-021-01153-z
Cao J et al (2017) Comprehensive single-cell transcriptional profiling of a multicellular organism. Science (New York, N.Y.) 357:661–667. https://doi.org/10.1126/science.aam8940
Cao J et al (2019) The single-cell transcriptional landscape of mammalian organogenesis. Nature 566:496–502. https://doi.org/10.1038/s41586-019-0969-x
Rosenberg AB et al (2018) Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science (New York, N.Y.) 360:176–182. https://doi.org/10.1126/science.aam8999
Shashikant T, Ettensohn CA (2019) Genome-wide analysis of chromatin accessibility using ATAC-seq. Methods Cell Biol 151:219–235. https://doi.org/10.1016/bs.mcb.2018.11.002
Yan F et al (2020) From reads to insight: a hitchhiker’s guide to ATAC-seq data analysis. Genome Biol 21:22. https://doi.org/10.1186/s13059-020-1929-3
Cusanovich DA et al (2015) Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science (New York, N.Y.) 348:910–914. https://doi.org/10.1126/science.aab1601
Lareau CA et al (2019) Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat Biotechnol 37:916–924. https://doi.org/10.1038/s41587-019-0147-6
Wang K et al (2021) Simple oligonucleotide-based multiplexing of single-cell chromatin accessibility. Mol Cell 81:4319-4332.e4310. https://doi.org/10.1016/j.molcel.2021.09.026
Ng PC, Kirkness EF (2010) Whole genome sequencing. Methods Mol Biol (Clifton, N.J.) 628:215–226. https://doi.org/10.1007/978-1-60327-367-1_12
Vitak SA et al (2017) Sequencing thousands of single-cell genomes with combinatorial indexing. Nat Methods 14:302–308. https://doi.org/10.1038/nmeth.4154
Moore LD et al (2013) DNA methylation and its basic function. Neuropsychopharmacology 38:23–38. https://doi.org/10.1038/npp.2012.112
Gouil Q, Keniry A (2019) Latest techniques to study DNA methylation. Essays Biochem 63:639–648. https://doi.org/10.1042/ebc20190027
Frommer M et al (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA 89:1827–1831. https://doi.org/10.1073/pnas.89.5.1827
Lister R et al (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322. https://doi.org/10.1038/nature08514
Smallwood SA et al (2014) Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 11:817–820. https://doi.org/10.1038/nmeth.3035
Farlik M et al (2015) Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep 10:1386–1397. https://doi.org/10.1016/j.celrep.2015.02.001
Farlik M et al (2016) DNA methylation dynamics of human hematopoietic stem cell differentiation. Cell Stem Cell 19:808–822. https://doi.org/10.1016/j.stem.2016.10.019
Angermueller C et al (2016) Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods 13:229–232. https://doi.org/10.1038/nmeth.3728
Clark SJ et al (2017) Genome-wide base-resolution mapping of DNA methylation in single cells using single-cell bisulfite sequencing (scBS-seq). Nat Protoc 12:534–547. https://doi.org/10.1038/nprot.2016.187
Luo C et al (2017) Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science (New York, N.Y.) 357:600–604. https://doi.org/10.1126/science.aan3351
Mulqueen RM et al (2018) Highly scalable generation of DNA methylation profiles in single cells. Nat Biotechnol 36:428–431. https://doi.org/10.1038/nbt.4112
Lieberman-Aiden E et al (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (New York, N.Y.) 326:289–293. https://doi.org/10.1126/science.1181369
Ramani V et al (2016) Understanding spatial genome organization: methods and insights. Genomics Proteomics Bioinformatics 14:7–20. https://doi.org/10.1016/j.gpb.2016.01.002
Eagen KP (2018) Principles of chromosome architecture revealed by Hi-C. Trends Biochem Sci 43:469–478. https://doi.org/10.1016/j.tibs.2018.03.006
Kong S, Zhang Y (2019) Deciphering Hi-C: from 3D genome to function. Cell Biol Toxicol 35:15–32. https://doi.org/10.1007/s10565-018-09456-2
Ramani V et al (2017) Massively multiplex single-cell Hi-C. Nat Methods 14:263–266. https://doi.org/10.1038/nmeth.4155
Hasin Y et al (2017) Multi-omics approaches to disease. Genome Biol 18:83. https://doi.org/10.1186/s13059-017-1215-1
Chappell L et al (2018) Single-sell (Multi)omics technologies. Annu Rev Genomics Hum Genet 19:15–41. https://doi.org/10.1146/annurev-genom-091416-035324
Mimitou EP et al (2019) Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat Methods 16:409–412. https://doi.org/10.1038/s41592-019-0392-0
Cao J et al (2018) Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science (New York, N.Y.) 361:1380–1385. https://doi.org/10.1126/science.aau0730
Mimitou EP et al (2021) Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat Biotechnol 39:1246–1258. https://doi.org/10.1038/s41587-021-00927-2
Swanson E et al (2021) Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq. Elife. https://doi.org/10.7554/eLife.63632
Hartmann FJ et al (2018) A universal live cell barcoding-platform for multiplexed human single cell analysis. Sci Rep 8(1):10770. https://doi.org/10.1038/s41598-018-28791-2
Mylka V et al (2022) Comparative analysis of antibody- and lipid-based multiplexing methods for single-cell RNA-seq. Genome Biol 23(1):55. https://doi.org/10.1186/s13059-022-02628-8
Cheng J et al (2021) Multiplexing methods for simultaneous large-scale transcriptomic profiling of samples at single-cell resolution. Adv Sci (Weinheim, Baden-Wurttemberg, Germany) 8:e2101229. https://doi.org/10.1002/advs.202101229
Rodriques SG et al (2019) Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363(6434):1463–1467. https://doi.org/10.1126/science.aaw1219
Vickovic S et al (2019) High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods 16(10):987–990. https://doi.org/10.1038/s41592-019-0548-y
Liu Y et al (2020) High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183(6):1665-1681.e18. https://doi.org/10.1016/j.cell.2020.10.026
Kaya-Okur HS et al (2019) CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10:1930. https://doi.org/10.1038/s41467-019-09982-5
Acknowledgements
We thank Kai He from Southern Medical University, and Yanfang Lu from Henan Provincial People’s Hospital for suggestion in the preparation of this manuscript.
Funding
This work was supported by the grants from the Natural Science Foundation of Guangdong Province (Major Basic Cultivation Project 2018B030308004, and Major Projects of Basic and Applied Basic Research 2019B1515120033), the National Nature Science Foundation of China (32071452), the Open Fund Programs of Shenzhen Bay Laboratory (SZBL2020090501003), and the Pearl River Talents Program Local Innovative and Research Teams (2017BT01S131).
Author information
Authors and Affiliations
Contributions
All authors contributed to the review conception and design. Material preparation, data collection, and analysis were performed by YZ, SX, ZW, and JG. The first draft of the manuscript was written by YZ and SX, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest to declare.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Xu, S., Wen, Z. et al. Sample-multiplexing approaches for single-cell sequencing. Cell. Mol. Life Sci. 79, 466 (2022). https://doi.org/10.1007/s00018-022-04482-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04482-0