Abstract
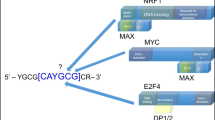
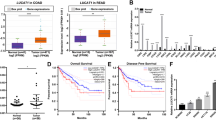
In addition to genomic alterations, aberrant changes in post-transcriptional regulation can modify gene function and drive cancer development. RNA-binding proteins (RBPs) are a large class of post-transcriptional regulators that have been increasingly implicated in carcinogenesis. By integrating multi-omics data, we identify LARP1 as one of the most upregulated RBPs in colorectal cancer (CRC) and demonstrate its oncogenic properties. We perform LARP1:RNA interactome profiling and unveil a previously unexplored role for LARP1 in targeting the 3′UTR of oncogenes in CRC. Notably, we identify the proto-oncogenic transcription factor MYC as a key LARP1-regulated target. Our data show that LARP1 positively modulates MYC expression by associating with its 3′UTR. In addition, antisense oligonucleotide-mediated blocking of the interaction between LARP1 and the MYC 3′UTR reduces MYC expression and in vitro CRC growth. Furthermore, a systematic analysis of LARP1:protein interactions reveals IGF2BP3 and YBX1 as LARP1-interacting proteins that also regulate MYC expression and CRC development. Finally, we demonstrate that MYC reciprocally modulates LARP1 expression by targeting its enhancer. In summary, our data reveal a critical, previously uncharacterized role of LARP1 in promoting CRC tumorigenesis, validate its direct regulation of the proto-oncogene MYC and delineate a model of the positive feedback loop between MYC and LARP1 that promotes CRC growth and development.







Similar content being viewed by others
Availability of data and material
The authors confirmed that the data supporting the findings of this study are available within the article and its supplementary material.
References
Wurth L (2012) Versatility of RNA-binding proteins in cancer. Comp Funct Genomics 2012:178525
Hentze MW et al (2018) A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 19(5):327–341
Audic Y, Hartley RS (2004) Post-transcriptional regulation in cancer. Biol Cell 96(7):479–498
Arnold M et al (2017) Global patterns and trends in colorectal cancer incidence and mortality. Gut 66(4):683–691
Kuipers EJ et al (2015) Colorectal cancer. Nat Rev Dis Primers 1:15065
Xie C et al (2013) LARP1 predict the prognosis for early-stage and AFP-normal hepatocellular carcinoma. J Transl Med 11:272
Bousquet-Antonelli C, Deragon JM (2009) A comprehensive analysis of the La-motif protein superfamily. RNA 15(5):750–764
Hopkins TG et al (2016) The RNA-binding protein LARP1 is a post-transcriptional regulator of survival and tumorigenesis in ovarian cancer. Nucleic Acids Res 44(3):1227–1246
Mura M et al (2015) LARP1 post-transcriptionally regulates mTOR and contributes to cancer progression. Oncogene 34(39):5025–5036
Ye L et al (2016) Overexpression of LARP1 predicts poor prognosis of colorectal cancer and is expected to be a potential therapeutic target. Tumour Biol 37(11):14585–14594
Fonseca BD et al (2015) La-related Protein 1 (LARP1) represses terminal Oligopyrimidine (TOP) mRNA translation downstream of mTOR Complex 1 (mTORC1). J Biol Chem 290(26):15996–16020
Tcherkezian J et al (2014) Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5’TOP mRNA translation. Genes Dev 28(4):357–371
Butter F et al (2009) Unbiased RNA–protein interaction screen by quantitative proteomics. Proc Natl Acad Sci USA 106(26):10626–10631
Yoon JH, Srikantan S, Gorospe M (2012) MS2-TRAP (MS2-tagged RNA affinity purification): tagging RNA to identify associated miRNAs. Methods 58(2):81–87
Meisenheimer KM, Koch TH (1997) Photocross-linking of nucleic acids to associated proteins. Crit Rev Biochem Mol Biol 32(2):101–140
Van Nostrand EL et al (2016) Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13(6):508–514
Lovci MT et al (2013) Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol 20(12):1434–1442
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842
Li Q, Brown JB, Huang H, Bickel PJ (2011) Measuring reproducibility of high throughput experiments. Ann Appl Stat 5:1752–1779
Liao Y et al (2019) WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47(W1):W199–W205
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26(12):1367–1372
Lee TI et al (2006) Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protocol 1(2):729–748
Zhang B et al (2020) A comprehensive expression landscape of RNA-binding proteins (RBPs) across 16 human cancer types. RNA Biol 17(2):211–226
Lahr RM et al (2015) The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5’TOP sequence. Nucleic Acids Res 43(16):8077–8088
Lahr RM et al (2017) La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife. https://doi.org/10.7554/eLife.24146
Roy HK et al (2002) AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis 23(1):201–205
Rochlitz CF, Herrmann R, de Kant E (1996) Overexpression and amplification of c-myc during progression of human colorectal cancer. Oncology 53(6):448–454
Erisman MD et al (1985) Deregulation of c-myc gene expression in human colon carcinoma is not accompanied by amplification or rearrangement of the gene. Mol Cell Biol 5(8):1969–1976
Wang H, Birkenbach M, Hart J (2000) Expression of Jun family members in human colorectal adenocarcinoma. Carcinogenesis 21(7):1313–1317
Arber N et al (1996) Increased expression of cyclin D1 is an early event in multistage colorectal carcinogenesis. Gastroenterology 110(3):669–674
Sebolt-Leopold JS et al (1999) Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat Med 5(7):810–816
Cancer Genome Atlas N (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487(7407):330–337
Coller HA et al (2000) Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA 97(7):3260–3265
Smith DR, Myint T, Goh HS (1993) Over-expression of the c-myc proto-oncogene in colorectal carcinoma. Br J Cancer 68(2):407–413
Mayr C, Bartel DP (2009) Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138(4):673–684
Andres SF et al (2019) IMP1 3’ UTR shortening enhances metastatic burden in colorectal cancer. Carcinogenesis 40(4):569–579
Li Z et al (2012) An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev Cell 23(6):1176–1188
Stavraka C, Blagden S (2015) The la-related proteins, a family with connections to cancer. Biomolecules 5(4):2701–2722
Kim A et al (2020) Inhibition of Y box binding protein 1 suppresses cell growth and motility in colorectal cancer. Mol Cancer Ther 19(2):479–489
Xu W et al (2019) Increased IGF2BP3 expression promotes the aggressive phenotypes of colorectal cancer cells in vitro and vivo. J Cell Physiol 234(10):18466–18479
Bommert KS et al (2013) The feed-forward loop between YB-1 and MYC is essential for multiple myeloma cell survival. Leukemia 27(2):441–450
Palanichamy JK et al (2016) RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J Clin Invest 126(4):1495–1511
Dang CV (2012) MYC on the path to cancer. Cell 149(1):22–35
Berman AJ et al (2021) Controversies around the function of LARP1. RNA Biol 18(2):207–217
Chatel-Chaix L et al (2013) A host YB-1 ribonucleoprotein complex is hijacked by hepatitis C virus for the control of NS3-dependent particle production. J Virol 87(21):11704–11720
Wang Y et al (2015) C1QBP negatively regulates the activation of oncoprotein YBX1 in the renal cell carcinoma as revealed by interactomics analysis. J Proteome Res 14(2):804–813
Dejure FR et al (2017) The MYC mRNA 3’-UTR couples RNA polymerase II function to glutamine and ribonucleotide levels. EMBO J 36(13):1854–1868
Acknowledgements
We thank all past and present YT lab members for their constructive feedback on this project. We also thank Teck Kwang Lim from Protein and Proteomics Centre at the Department of Biological Sciences (DBS), NUS, and Charlene Chan from Cancer Science Institute of Singapore, for their assistance in performing the mass spectrometry analyses. In addition, we thank Avencia Sanchez-Mejias and Xiao Hong Chew for their technical assistance. Furthermore, we thank Shi Hao Tan from Cancer Science Institute of Singapore for his assistance in the design of the ChIP-qPCR experiments.
Funding
Y.T. is funded by a Singapore National Research Foundation Fellowship, a National Medical Research Council Open Fund—Individual Research Grant and a National University of Singapore President’s Assistant Professorship. Y.T., K.K. and D.K. are supported by the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education’s AcRF Tier 3 Grant (MOE2014-T3-1-006). G.W.Y. is funded by the National University of Singapore and National Research Foundation of Singapore. N.D. is supported by the NUS Research Scholarship. QYT is supported by a Cancer Science Institute of Singapore Research Scholarship. This research is supported by the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative, as well as the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education’s AcRF Tier 3 Grants, Grant number MOE2014-T3-1-006.
Author information
Authors and Affiliations
Contributions
YT, ND, and JJC conceptualized and designed the project. ND and QYT designed and performed experiments, analyzed and interpreted data and generated figures. VT, JJC, BZ, HT, ZHK, and CYL designed and performed experiments and analyzed data. HQT, KK, WJ, and GWY performed, analyzed and interpreted the eCLIP experiments. BZ and HY performed bioinformatics analysis and interpretation. CYL, SW, PCLL, BES, KCL, CSC, and KKT obtained and processed the patient samples. HTT, MCMC and DK performed mass spectrometry experiments and analysis. ND, QYT, JJC, YT, KK, BZ, HTT, and DK wrote and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Desi, N., Tong, Q.Y., Teh, V. et al. Global analysis of RNA-binding proteins identifies a positive feedback loop between LARP1 and MYC that promotes tumorigenesis. Cell. Mol. Life Sci. 79, 147 (2022). https://doi.org/10.1007/s00018-021-04093-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-021-04093-1