Abstract
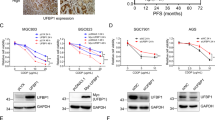
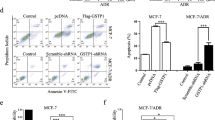
Glutathione S-transferase pi (GSTpi) is an important phase II detoxifying enzyme that participates in various physiological processes, such as antioxidant, detoxification, and signal transduction. The high expression level of GSTpi has been reported to be related to drug-resistant and anti-inflammatory and it functioned via its non-catalytic ligandin. However, the previous protection mechanism of GSTpi in DNA damage has not been addressed so far. Nijmegen breakage syndrome 1 (NBS1) is one of the most important sensor proteins to detect damaged DNA. Here, we investigated the interaction between GSTpi and NBS1 in HEK-293 T cells and human breast adenocarcinoma cells during DNA damage. Our results showed that overexpression of GSTpi in cells by transfecting DNA vector decreased the DNA damage level after methyl methanesulfonate (MMS) or adriamycin (ADR) treatment. We found that cytosolic GSTpi could increase NBS1 ubiquitin-mediated degradation in unstimulated cells, which suggested that GSTpi could maintain the basal level of NBS1 during normal conditions. In response to DNA damage, GSTpi can be phosphorylated in Ser184 and inhibit the ubiquitination degradation of NBS1 mediated by Skp2 to recover NBS1 protein level. Phosphorylated GSTpi can further enhance NBS1 nuclear translocation to activate the ATM-Chk2-p53 signaling pathway. Finally, GSTpi blocked the cell cycle in the G2/M phase to allow more time for DNA damage repair. Thus, our finding revealed the novel mechanism of GSTpi via its Ser184 phosphorylation to protect cells from cell death during DNA damage and it enriches the function of GSTpi in drug resistance.









Similar content being viewed by others
Availability of data and materials
Not applicable.
Code availability
Not applicable.
References
Chatterjee N, Walker GC (2017) Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen 58:235–263. https://doi.org/10.1002/em.22087
Townsend DM, Tew KD (2003) The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene 22:7369–7375. https://doi.org/10.1038/sj.onc.1206940
Tew KD, Manevich Y, Grek C, Xiong Y, Uys J, Townsend DM (2011) The role of glutathione S-transferase P in signaling pathways and S-glutathionylation in cancer. Free Radic Biol Med 51:299–313. https://doi.org/10.1016/j.freeradbiomed.2011.04.013
Josephy PD (2010) Genetic variations in human glutathione transferase enzymes: significance for pharmacology and toxicology. Hum Genomics Proteomics 2010:876940. https://doi.org/10.4061/2010/876940
Tew KD (2016) Glutathione-associated enzymes in anticancer drug resistance. Cancer Res 76:7–9. https://doi.org/10.1158/0008-5472.CAN-15-3143
Ciaccio PJ, Tew KD, LaCreta FP (1991) Enzymatic conjugation of chlorambucil with glutathione by human glutathione S-transferases and inhibition by ethacrynic acid. Biochem Pharmacol 42:1504–1507. https://doi.org/10.1016/0006-2952(91)90468-k
Adler V, Yin Z, Fuchs SY, Benezra M, Rosario L, Tew KD, Pincus MR, Sardana M, Henderson CJ, Wolf CR et al (1999) Regulation of JNK signaling by GSTp. Embo J 18:1321–1334. https://doi.org/10.1093/emboj/18.5.1321
Wu Y, Fan Y, Xue B, Luo L, Shen J, Zhang S, Jiang Y, Yin Z (2006) Human glutathione S-transferase P1–1 interacts with TRAF2 and regulates TRAF2-ASK1 signals. Oncogene 25:5787–5800. https://doi.org/10.1038/sj.onc.1209576
Chen D, Liu J, Rui B, Gao M, Zhao N, Sun S, Bi A, Yang T, Guo Y, Yin Z, Luo L (2014) GSTpi protects against angiotensin II-induced proliferation and migration of vascular smooth muscle cells by preventing signal transducer and activator of transcription 3 activation. Biochim Biophys Acta 1843:454–463. https://doi.org/10.1016/j.bbamcr.2013.11.024
Dong X, Yang Y, Zhou Y, Bi X, Zhao N, Zhang Z, Li L, Hang Q, Zhang R, Chen D et al (2019) Glutathione S-transferases P1 protects breast cancer cell from adriamycin-induced cell death through promoting autophagy. Cell Death Differ 26:2086–2099. https://doi.org/10.1038/s41418-019-0276-y
Kamada K, Goto S, Okunaga T, Ihara Y, Tsuji K, Kawai Y, Uchida K, Osawa T, Matsuo T, Nagata I, Kondo T (2004) Nuclear glutathione S-transferase pi prevents apoptosis by reducing the oxidative stress-induced formation of exocyclic DNA products. Free Radic Biol Med 37:1875–1884. https://doi.org/10.1016/j.freeradbiomed.2004.09.002
Paull TT (2015) Mechanisms of ATM Activation. Annu Rev Biochem 84:711–738. https://doi.org/10.1146/annurev-biochem-060614-034335
Choi M, Kipps T, Kurzrock R (2016) ATM mutations in cancer: therapeutic implications. Mol Cancer Ther 15:1781–1791. https://doi.org/10.1158/1535-7163.Mct-15-0945
Lee JH, Paull TT (2007) Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26:7741–7748. https://doi.org/10.1038/sj.onc.1210872
Wang Z, Gong Y, Peng B, Shi R, Fan D, Zhao H, Zhu M, Zhang H, Lou Z, Zhou J et al (2019) MRE11 UFMylation promotes ATM activation. Nucleic Acids Res 47:4124–4135. https://doi.org/10.1093/nar/gkz110
Sun Y, McCorvie TJ, Yates LA, Zhang X (2020) Structural basis of homologous recombination. Cell Mol Life Sci 77:3–18. https://doi.org/10.1007/s00018-019-03365-1
Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR 3rd, Hays L, Morgan WF, Petrini JH (1998) The hMre11/hRad50 protein complex and nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477–486. https://doi.org/10.1016/s0092-8674(00)81175-7
Taylor AM, Groom A, Byrd PJ (2004) Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair (Amst) 3:1219–1225. https://doi.org/10.1016/j.dnarep.2004.04.009
Goto S, Ihara Y, Urata Y, Izumi S, Abe K, Koji T, Kondo T (2001) Doxorubicin-induced DNA intercalation and scavenging by nuclear glutathione S-transferase pi. Faseb J 15:2702–2714. https://doi.org/10.1096/fj.01-0376com
Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y et al (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160–1166. https://doi.org/10.1126/science.1140321
Blackford AN, Jackson SP (2017) ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell 66:801–817. https://doi.org/10.1016/j.molcel.2017.05.015
Manic G, Obrist F, Sistigu A, Vitale I (2015) Trial watch: targeting ATM-CHK2 and ATR-CHK1 pathways for anticancer therapy. Mol Cell Oncol 2:e1012976. https://doi.org/10.1080/23723556.2015.1012976
Luo Q, Guo H, Kuang P, Cui H, Deng H, Liu H, Lu Y, Wei Q, Chen L, Fang J et al (2018) Sodium fluoride arrests renal G2/M phase cell-cycle progression by activating ATM-Chk2-P53/Cdc25C signaling pathway in mice. Cell Physiol Biochem 51:2421–2433. https://doi.org/10.1159/000495899
Zhou Y, Cao X, Yang Y, Wang J, Yang W, Ben P, Shen L, Cao P, Luo L, Yin Z (2018) Glutathione S-transferase Pi prevents sepsis-related high mobility group box-1 protein translocation and release. Front Immunol 9:268. https://doi.org/10.3389/fimmu.2018.00268
Benedict B, van Harn T, Dekker M, Hermsen S, Kucukosmanoglu A, Pieters W, Delzenne-Goette E, Dorsman JC, Petermann E, Foijer F, Te Riele H (2018) Loss of p53 suppresses replication-stress-induced DNA breakage in G1/S checkpoint deficient cells. Elife. https://doi.org/10.7554/eLife.37868
Sun P, Wu H, Huang J, Xu Y, Yang F, Zhang Q, Xu X (2018) Porcine epidemic diarrhea virus through p53-dependent pathway causes cell cycle arrest in the G0/G1 phase. Virus Res 253:1–11. https://doi.org/10.1016/j.virusres.2018.05.019
Lee CC, Lin ML, Meng M, Chen SS (2018) Galangin induces p53-independent S-phase arrest and apoptosis in human nasopharyngeal carcinoma cells through inhibiting PI3K-AKT signaling pathway. Anticancer Res 38:1377–1389. https://doi.org/10.21873/anticanres.12361
Lavin MF, Kozlov S, Gatei M, Kijas AW (2015) ATM-dependent phosphorylation of all three members of the MRN complex: from sensor to adaptor. Biomolecules 5:2877–2902. https://doi.org/10.3390/biom5042877
Ha GH, Ji JH, Chae S, Park J, Kim S, Lee JK, Kim Y, Min S, Park JM, Kang TH et al (2019) Pellino1 regulates reversible ATM activation via NBS1 ubiquitination at DNA double-strand breaks. Nat Commun 10:1577. https://doi.org/10.1038/s41467-019-09641-9
Tseng SF, Chang CY, Wu KJ, Teng SC (2005) Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J Biol Chem 280:39594–39600. https://doi.org/10.1074/jbc.M508425200
Wu J, Zhang X, Zhang L, Wu CY, Rezaeian AH, Chan CH, Li JM, Wang J, Gao Y, Han F et al (2012) Skp2 E3 ligase integrates ATM activation and homologous recombination repair by ubiquitinating NBS1. Mol Cell 46:351–361. https://doi.org/10.1016/j.molcel.2012.02.018
Shi H, Lu D, Shu Y, Shi W, Lu S, Wang K (2008) Expression of multidrug-resistance-related proteins P-glycoprotein, glutathione-S-transferases, topoisomerase-II and lung resistance protein in primary gastric cardiac adenocarcinoma. Cancer Invest 26:344–351. https://doi.org/10.1080/07357900701788072
Yu ST, Chen TM, Chern JW, Tseng SY, Chen YH (2009) Downregulation of GSTpi expression by tryptanthrin contributing to sensitization of doxorubicin-resistant MCF-7 cells through c-jun NH2-terminal kinase-mediated apoptosis. Anticancer Drugs 20:382–388. https://doi.org/10.1097/CAD.0b013e32832a2cd4
Basu AK (2018) DNA damage, mutagenesis and cancer. Int J Mol Sci. https://doi.org/10.3390/ijms19040970
Li Z, Pearlman AH, Hsieh P (2016) DNA mismatch repair and the DNA damage response. DNA Repair (Amst) 38:94–101. https://doi.org/10.1016/j.dnarep.2015.11.019
Roos WP, Thomas AD, Kaina B (2016) DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 16:20–33. https://doi.org/10.1038/nrc.2015.2
O’Connor MJ (2015) Targeting the DNA damage response in cancer. Mol Cell 60:547–560. https://doi.org/10.1016/j.molcel.2015.10.040
Singh S, Okamura T, Ali-Osman F (2010) Serine phosphorylation of glutathione S-transferase P1 (GSTP1) by PKCalpha enhances GSTP1-dependent cisplatin metabolism and resistance in human glioma cells. Biochem Pharmacol 80:1343–1355. https://doi.org/10.1016/j.bcp.2010.07.019
Theodossiou TA, Olsen CE, Jonsson M, Kubin A, Hothersall JS, Berg K (2017) The diverse roles of glutathione-associated cell resistance against hypericin photodynamic therapy. Redox Biol 12:191–197. https://doi.org/10.1016/j.redox.2017.02.018
Steinkellner H, Hoelzl C, Uhl M, Cavin C, Haidinger G, Gsur A, Schmid R, Kundi M, Bichler J, Knasmüller S (2005) Coffee consumption induces GSTP in plasma and protects lymphocytes against (+/-)-anti-benzo [a]pyrene-7,8-dihydrodiol-9,10-epoxide induced DNA-damage: results of controlled human intervention trials. Mutat Res 591:264–275. https://doi.org/10.1016/j.mrfmmm.2005.04.016
Feng J, Islam A, Bean B, Feng J, Sparapani S, Shrivastava M, Goyal A, Omran RP, Mallick J, Whiteway M (2020) Hof1 plays a checkpoint-related role in MMS-induced DNA damage response in Candida albicans. Mol Biol Cell 31:348–359. https://doi.org/10.1091/mbc.E19-06-0316
Kumar K, Moirangthem R, Kaur R (2020) Histone H4 dosage modulates DNA damage response in the pathogenic yeast Candida glabrata via homologous recombination pathway. PLoS Genet 16:e1008620. https://doi.org/10.1371/journal.pgen.1008620
Wu B, Dong D (2012) Human cytosolic glutathione transferases: structure, function, and drug discovery. Trends Pharmacol Sci 33:656–668. https://doi.org/10.1016/j.tips.2012.09.007
Bartolini D, Galli F (2016) The functional interactome of GSTP: A regulatory biomolecular network at the interface with the Nrf2 adaption response to oxidative stress. J Chromatogr B Analyt Technol Biomed Life Sci 1019:29–44. https://doi.org/10.1016/j.jchromb.2016.02.002
Yan S, Sorrell M, Berman Z (2014) Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell Mol Life Sci 71:3951–3967. https://doi.org/10.1007/s00018-014-1666-4
Medunjanin S, Putzier M, Nöthen T, Weinert S, Kähne T, Luani B, Zuschratter W, Braun-Dullaeus RC (2020) DNA-PK: gatekeeper for IKKγ/NEMO nucleocytoplasmic shuttling in genotoxic stress-induced NF-kappaB activation. Cell Mol Life Sci 77:4133–4142. https://doi.org/10.1007/s00018-019-03411-y
Lavin MF (2008) Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 9:759–769. https://doi.org/10.1038/nrm2514
Bartek J, Lukas J (2003) Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3:421–429. https://doi.org/10.1016/s1535-6108(03)00110-7
Song S, Shi Y, Wu W, Wu H, Chang L, Peng P, Zhang L, Fan J, Gu J, Ruan Y (2020) Reticulon 3-mediated Chk2/p53 activation suppresses hepatocellular carcinogenesis and is blocked by hepatitis B virus. Gut. https://doi.org/10.1136/gutjnl-2020-321386
LeBron C, Chen L, Gilkes DM, Chen J (2006) Regulation of MDMX nuclear import and degradation by Chk2 and 14–3-3. Embo j 25:1196–1206. https://doi.org/10.1038/sj.emboj.7601032
Ertych N, Stolz A, Valerius O, Braus GH, Bastians H (2016) CHK2-BRCA1 tumor-suppressor axis restrains oncogenic aurora-A kinase to ensure proper mitotic microtubule assembly. Proc Natl Acad Sci USA 113:1817–1822. https://doi.org/10.1073/pnas.1525129113
Lee JH, Paull TT (2005) ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308:551–554. https://doi.org/10.1126/science.1108297
Falck J, Coates J, Jackson SP (2005) Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434:605–611. https://doi.org/10.1038/nature03442
Xu Y, Wu W, Han Q, Wang Y, Li C, Zhang P, Xu H (2019) Post-translational modification control of RNA-binding protein hnRNPK function. Open Biol 9:180239. https://doi.org/10.1098/rsob.180239
Acknowledgements
We thank Prof. Zhigang Guo (China) for sharing MCF-7/ADR cells.
Funding
This work was financially supported by grants from the Natural Science Foundation of China (32001023, 81671565, 81771703 and 31901012), the China Postdoctoral Science Foundation (2020T130058ZX), and the Priority Academic Program Development of Jiangsu Higher Education Institution (PAPD).
Author information
Authors and Affiliations
Contributions
Conceptualization: ZY, LL, PC, and JZ; methodology: ZY, LL, PC, and JZ; formal analysis: ZY, LL, PC, and JZ; investigation: JZ, LG, YS, and TH; validation: XF, XB, SL, and JL; writing—original draft preparation: ZY and JZ; writing—review and editing: ZY; funding acquisition: ZY, XB, LL, and LG; resources: SL and LG; visualization: LL and PC; supervision: ZY, LL, and PC.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Ethics approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
18_2021_4057_MOESM1_ESM.tif
Supplementary figure 1 (TIF 13275 KB) Fig. S1 GSTpi activates ATM-Chk2-p53 signal pathway during DNA damage. a HEK-293T-GSTpi-/- cells transfected with pcDNA3.1 (2 μg/mL) or Flag-GSTpi (2 μg/mL) were treated with 0.5 mM MMS or not for 1 h, then cells were allowed to recover in normal medium for 6 h, then cell lysates were subjected to immunoblotting using the indicated antibodies. Data shown are representative of three independent experiments. b HEK-293T-WT cells transfected with pLKO.1 (2 μg/mL) or GSTpi-shRNA (2 μg/mL) were treated with 0.5 mM MMS or not for 1 h, then cells were allowed to recover in normal medium for 6 h, then cell lysates were subjected to immunoblotting using the indicated antibodies. Data shown are representative of three independent experiments. c HEK-293T-WT cells transfected with pcDNA3.1 (2 μg/mL) or Flag-GSTpi (2 μg/mL) were pre-incubated with Ku55933 (10 μM) for 4 h, then were treated with MMS (0.5 mM) for 1 h, cells were allowed to recover in medium containing Ku55933 (10 μM) for 6 h, then cell lysates were subjected to immunoblotting using the indicated antibodies. Data represent the mean ± SD of at least three independent experiments. Statistics were calculated by unpaired Student’s t test. ** p < 0.01. d HEK-293T-WT cells transfected with pcDNA3.1 (2 μg/mL) or Flag-GSTpi (2 μg/mL) were pre-incubated with Pifithrin-α (10 μM) for 2 h, then were treated with MMS (0.5 mM) for 1 h, cells were allowed to recover in medium containing Pifithrin-α (10 μM) for 6 h, then cell lysates were subjected to immunoblotting using the indicated antibodies. Data represent the mean ± SD of at least three independent experiments. Statistics were calculated by unpaired Student’s t test. * p < 0.05, ** p < 0.01
18_2021_4057_MOESM2_ESM.tif
Supplementary figure 2 (TIF 3970 KB) Fig. S2 GSTpi regulates NBS1 in a transferase activity dependent way. a HEK-293T-GSTpi-/- cells transfected with pcDNA3.1 (2 μg/mL) or Flag-GSTpi (2 μg/mL) were treated with 0.5 mM MMS or not for 1 h, then cells were allowed to recover in normal medium for 6 h, and cell lysates were subjected to immunoblotting using the indicated antibodies. Data represent the mean ± SD of at least three independent experiments. Statistics were calculated by unpaired Student’s t test. * p < 0.05, ** p < 0.01, *** p < 0.001
18_2021_4057_MOESM3_ESM.tif
Supplementary figure 3 (TIF 4232 KB) Fig. S3 Phosphorylated GSTP by cPKC involved in NBS1 nuclear translocation. a HEK-293T-GSTpi-/- cells transfected with pcDNA3.1 (2 μg/mL), Flag-GSTpi (2 μg/mL), GSTpi-S184A (2 μg/mL) or GSTpi-S184D (2 μg/mL) were treated with 100 nM PMA or not for 1 h, after which the extracted nuclear and cytoplasmic fractions were subjected to Western blot analysis for NBS1 and Flag. Data represent the mean ± SD of at least three independent experiments. Statistics were calculated by unpaired Student’s t test. * p < 0.05, ** p < 0.01, n.s.: not significant (p > 0.05)
Rights and permissions
About this article
Cite this article
Zhou, J., Gu, L., Shi, Y. et al. GSTpi reduces DNA damage and cell death by regulating the ubiquitination and nuclear translocation of NBS1. Cell. Mol. Life Sci. 79, 54 (2022). https://doi.org/10.1007/s00018-021-04057-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-021-04057-5