Abstract
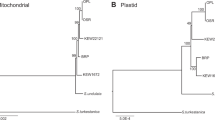
The mitochondrial DNA of plant and animal cells is a transcriptionally active genome that traces its origins to a symbiotic infection of eucaryotic cells by bacterial progenitors. As prescribed by the Serial Endosymbiosis Theory, symbiotic organelles have gradually transferred their genes to the eucaryotic genome, producing a functional interaction of nuclear and mitochondrial genes in organelle function. We report here a recent remarkable transposition of 7.9 kb of a typically 17.0-kb mitochondrial genome to a specific nuclear chromosomal position in the domestic cat. The integrated segment has subsequently become amplified 38–76 times and now occurs as a tandem repeat macrosatellite with multiple-length alleles resolved by pulse-field gel electrophoresis (PFGE) segregating in cat populations. Sequence determination of the nuclear mitochondrial DNA segment, Numt, revealed a d(CA)-rich 8-bp motif [ACACACGT] repeated imperfectly five times at the deletion junction that is a likely target for recombination. The extent and pattern of sequence divergence of Numt genes from the cytoplasmic mtDNA homologues plus the occurrence of Numt in other species of the family Felidae allowed an estimate for the origins of Numt at 1.8–2.0 million years ago in an ancestor of four modern species in the genus Felis. Numt genes do not function in cats; rather, the locus combines properties of nuclear minisatellites and pseudogenes. These observations provide an empirical glimpse of historic genomic events that may parallel the accommodation of organelles in eucaryotes.
Similar content being viewed by others
References
Anderson S, Bankier AT, Barrell BG, De Bruijn MHL, Coulson AR, Drouin J, Eperon DP, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290: 457–465
Anderson S, De Bruijn MHL, Coulson AR, Eperon IC, Sanger F, Young IG (1982) Complete sequence of bovine mitochondrial DNA. J Mol Biol 156: 683–717
Arnason U, Johnsson E (1992) The complete mitochondrial DNA sequence of the harbor seal mtDNA, Phoca vitulina. J Mol Evol 34: 493–505
Attardi G (1985) Animal mitochondrial DNA: an extreme example of genetic economy. Int Rev Cytol 93: 93–145
Avise JC (1991) Ten unorthodox perspectives on evolution prompted by comparative population genetic finding on mitochondrial DNA. Ann Rev Genet 25: 45–69
Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA (1981) Sequence and gene organization of mouse mitochondrial DNA. Cell 26: 167–180
Birky CW Jr (1978) Transmission genetics of mitochondria and chloroplasts. Ann Rev Genet 12: 471–512
Birren WB, Wai E, Hook L, Simon MI (1988) Optimized conditions for pulsed field gel electrophoresis separations of DNA. Nucleic Acids Res 16: 7563–7582
Breitenberger C, RajBhandary UL (1985) Some highlights of mitochondrial research based on analyses of Neurospora crassa mitochondrial DNA, Trends Biochem Sci 10: 478–483
Brown WM (1985) The mitochondrial genome of animals. In: MacIntyre RJ (ed) Molecular evolutionary genetics. Plenum Press, New York, pp 95–130
Capecchi MR (1989) Altering the genome by homologous recombination. Science 244: 1288–1292
Chang DD, Clayton DA (1984) Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell 36: 635–643
Collier GE, O'Brien SJ (1985) A molecular phylogeny of the Felidae: immunological distance. Evolution 39: 473–487
Devereux J, Haeberli P, Smithies O (1984) A comprehensive set of sequence analysis programs for the Vax. Nucleic Acids Res 12: 387–395
Dietrich W, Katz H, Lincoln SE, Shin H-S, Friedman J, Dracopoli NC, Lander ES (1992) A genetic map of the mouse suitable for typing intraspecific crosses. Genetics 131: 423–447
Drouin J (1980) Cloning of human mitochondrial DNA in Escherichia coli. J Mol Biol 140: 15–34
Dujon B, Belcour L (1989) Mitochondrial DNA instabilities. In Berg DE, Howe MM (eds) Mobile DNA. American Society Microbiology, Washington, DC, pp 861–878
Ellis J (1982) Promiscuous DNA-chloroplast genes inside plant mitochondria. Nature 299: 678–680
Eperon IC, Anderson S, Nierlich DP (1980) Distinctive sequence of human mitochondrial ribosomal RNA genes. Nature 286: 460–467
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17: 368–376
Felsenstein J (1993) PHYLIP: phylogenetic inference package, version 3.5c. University of Washington
Fitch WM, Margoliash E (1967) Construction of phylogenetic trees. Science 135: 279–284
Fukuda M, Wakasugi S, Tsuzuki T, Nomiyama H, Shimada K (1985) Mitochondrial DNA-like sequences in the human nuclear genome. J Mol Biol 186: 257–266
Gadaleta G, Pepe G, De Candia C, Quagliariello C, Sbisa E, Saccone C (1989) The complete nucleotide sequence of the (Rattus norvegicus mitochondria) genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol 28: 497–516
Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD (1991) Transfer of rp122 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J 10: 3073–3078
Gellissen G, Michaelis G (1987) Gene transfer: mitochondria to nucleus. In: Lee JJ, Frederick JF (eds) Endocytobiology. Ann NY Acad Sci 503: 391
Gilbert DA, O'Brien JS, O'Brien SJ (1988) Chromosomal mapping of lysosomal enzyme structural genes in the domestic cat. Genomics 2: 329–336
Gray MW (1989) The evolutionary origins of organelles. Trends Genet 5: 294–299
Gyllensten UB, Erlich HA (1988) Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci USA 85: 7652–7656
Hartl F-U, Neupert W (1990) Protein sorting to mitochondria: evolutionary conservations of folding and assembly. Science 247: 930–939
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22: 160–174
Hayashi JI, Tagashira Y, Yoshida MC (1985) Absence of extensive recombination between inter- and intraspecies mitochondrial DNA in mammalian cells. Exp Cell Res 160: 387–395
Hoehe MR, Caenazzo L, Martinez MM, Hsieh WT, Modi WS, Gershon ES, Bonner TI (1991) Genetic and physical mapping of the human cannabinoid receptor gene to chromosome 6g14-q15. New Biol 3: 880–885
Hoelzel AR, Hancock JM, Dover GA (1993) Generation of VNTRs and heteroplasmy by sequence turnover in the mitochondrial control region of two elephant seal species. J Mol Evol 37: 190–197
Hughes AL, Nei M (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335: 167–170
Innis MA, Gelfand DH, Srinsky JJ, White TJ (eds) (1990) PCR protocols. Academic Press, San Diego, CA
Jeffreys AJ, MacLead A, Tamaki K, Neil DL, Moncleton DG (1991) Minisatellite repeat coding as a digital approach to DNA typing. Nature 354: 204–209
Kamimura N, Ishii S, Linadong M, Shay JW (1989) Three separate mtDNA sequences are contiguous in human genomic DNA. J Mol Biol 210: 703–707
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge, England
Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanca FX, Wilson AC (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA 86: 6196–6200
Kurten B (1968) Pleistocene mammals of Europe. Aldine Press, Chicago
Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4: 203–221
Li WH, Gojobori T, Nei M (1981) Pseudogenes as a paradigm of neutral evolution. Nature 292: 237–239
Locker J, Lewin A, Rabinowitz M (1979) The structure and organization of mitochondrial DNA from petite yeast. Plasmid 2: 155–181
Margulis L (1970) Origin of eukaryotic cells. Yale University Press, New Haven, CT
Masuda R, Yuhki N, O'Brien SJ (1991) Molecular cloning, chromosomal assignment and nucleotide sequences of the feline homeobox HOX3A. Genomics 11: 1007–1013
Masuda R, Yuhki N, Lopez JV, O'Brien SJ (1994) A molecular phylogeny of the Felidae family, based on DNA sequences of mitochondrial 12S rRNA and cytochrome b genes (in preparation)
Mindell DP, Honeycutt RL (1990) Ribosomal RNA in vertebrates: evolution and phylogenetic applications. Ann Rev Ecol Syst 21: 541–566
Moritz C, Dowling TE, Brown WM (1987) Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Ann Rev Ecol Syst 18: 269–92
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nowak RM (1991) Walker's mammals of the world, 5th ed. The Johns Hopkins University Press, Baltimore, MD
Nugent JM, Palmer JD (1991) RNA-mediated transfer of the gene COXII from the mitochondrion to the nucleus during flowering plant evolution. Cell 66: 473–481
O'Brien SJ (1986) Molecular genetics in the domestic cat and its relatives. Trends Genet 2: 137–143
O'Brien SJ, Collier GE, Benveniste RE, Nash WG, Newman AK, Simonson JM, Eichelberger MA, Seal US, Janssen D, Bush M, Wildt DE (1987) Setting the molecular clock in Felidae: the great cats, Panthera. In: Tilson RL, Seal US (eds) Tigers of the world. Noyes Publications, Park Ridge, NJ, pp 10–27
O'Brien SJ, Nash WG (1982) Genetic mapping in mammals: chromosome map of domestic cat. Science 216: 257–265
O'Brien SJ, Roelke ME, Yuhki N, Richards KW, Johnson WE, Franklin WL, Anderson AE, Bass OL Jr, Belden RC, Martenson JS (1990) Genetic introgression within the Florida panther Felis concolor coryi. Natl Geo Res 6: 485–494
Ohta T (1992) The nearly neutral theory of molecular evolution. Ann Rev Ecol Syst 23: 263–286
Quigley F, Martin WF, Ceriff R (1988) Intron conservation across the prokaryotic-eucaryotic boundary: structure of the nuclear gene for chloroplast glyceraldehyde 3-phosphate dehydrogenase from maize. Proc Natl Acad Sci USA 85: 2672–2676
Saccone C, Pesole G, Sbisa E (1991) The main regulatory region of mammalian mitochondrial DNA: structure-function model and evolutionary pattern. J Mol Evol 33: 83–91
Saitou N, Nei M (1987) The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Schinkel AH, Tabak HF (1989) Mitochondrial RNA polymerase: dual role in transcription and replication. Trends Genet 5: 149–154
Schon EA, Rizzuto R, Moraes CT, Nakase H, Zeriani M, Dimauro S (1989) A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science 24: 346–349
Serikawa T, Kuramoto T, Hilbert P, Mori M, Yamada J, Dubay CJ, Lindpainter K, Ganten D, Guenet JL, Lathrop GM, Beckmann JS (1992) Rat gene mapping using PCR-analyzed microsatellites. Genetics 131: 701–702
Smith MF, Thomas WK, Patton JL (1991) Mitochondrial DNA-like sequence in the nuclear genome of an akodontine rodent. Mol Biol Evol 9: 204–215
Swofford DL (1990) Phylogenetic analysis using parsimony (PAUP), version 3.0. Illinois Natural History Survey, Champaign, IL
Tsuzuki T, Nomiyama H, Setoyaja C, Maeda S, Shimada K (1983) Presence of mitochondrial-DNA-like sequences in the human nuclear DNA. Gene 25: 223–229
Van den Boogaart P, Samalio J, Agsteribbe E (1982) Similar genes for a mitochondrial ATPase subunit in the nuclear and mitochondrial genomes of Neurospora crassa. Nature 298: 187–189
Wahls WP, Wallace LJ, Moore PD (1990) Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell 60: 95–103
Wakasugi S, Nomiyama H, Fukuda M, Tsuzuki T, Shimada K (1985) Insertion of a long Kpn I family member within a mitochondrial DNA-like sequence present in the human nuclear genome. Gene 36: 281–288
Wallace DC, Lott MT, Torroni A, Shoffner JM (1991) Report of the committee on human mitochondrial DNA. Cytogenet Cell Genet 58: 1103–1123
Weber JL (1990) Human DNA polymorphisms based on length variations in simple-sequence tandem repeats. Genome Analysis 1: 159–181
Wilson AC, Cann RL, Carr SM, George M, Gyllensten UB, Helm-Bychowski KM, Higuchi RG, Palumbi SR, Prager EM, Sage RD, Stoneking M (1985) Mitochondrial DNA and two perspectives on evolutionary genetics. Biol J Linnean Soc 26: 375–400
Woese CR (1987) Bacterial evolution. Micro Rev 51: 221–271
Wurster-Hill DH, Centerwall WR (1982) The interrelationships of chromosome patterns in canids, mustelids, hyena and felids. Cytogenet Cell Genet 34: 178–192
Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR (1985) Mitochondrial origins. Proc Natl Acad Sci USA 82: 4443–4447
Yuhki N, O'Brien SJ (1990) DNA recombination and natural selection pressure sustain genetic sequence diversity of the feline MHC class I genes. J Exp Med 172: 621–630
Zullo S, Sieu LC, Slightom JL, Hadler HI, Eisenstadt JM (1991) Mitochondrial D-loop sequences are integrated in the rat nuclear genome. J Mol Biol 221: 1223–1235
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Lopez, J.V., Yuhki, N., Masuda, R. et al. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol 39, 174–190 (1994). https://doi.org/10.1007/BF00163806
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00163806