Abstract
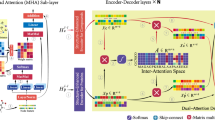
Drug-Target protein Interaction (DTI) prediction is a crucial task in the field of drug discovery. Prediction methods based on deep learning have been demonstrated to significantly enhance the accuracy of DTI prediction. Existing approaches mainly extract features from drug molecular sequences and then utilize networks for learning and prediction. However, drug molecular images can clearly display features such as atoms, structures, and chemical bonds, which are difficult to capture in sequences. Therefore, this study introduces a deep learning approach based on multi-view feature fusion, leveraging Transformer to combine the graph structure and image features of drug molecules, thereby learning more comprehensive drug features. This enables the model to learn more intricate interaction features between amino acids and atoms during DTI simulation. The proposed model was evaluated on three benchmark datasets and demonstrated significant improvements over the latest baselines. Furthermore, to validate the efficacy of capturing drug image feature information, ablation experiments were conducted, indicating a notable enhancement in accuracy upon incorporating image data.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Manoochehri, H.E., Nourani, M.: Drug-target interaction prediction using semi-bipartite graph model and deep learning. BMC Bioinformatics 21, 1–16 (2020)
Maia, E.H.B., Assis, L.C., De Oliveira, T.A., Da Silva, A.M., Taranto, A.G.: Structure-based virtual screening: from classical to artificial intelligence. Front. Chem. 8, 343 (2020)
Himmat, M., Salim, N., Al-Dabbagh, M.M., Saeed, F., Ahmed, A.: Adapting document similarity measures for ligand-based virtual screening. Molecules 21(4), 476 (2016)
Lee, I., Keum, J., Nam, H.: DeepConv-DTI: prediction of drug-target interactions via deep learning with convolution on protein sequences. PLoS Comput. Biol. 15(6), e1007129 (2019)
Öztürk, H., Özgür, A., Ozkirimli, E.: DeepDTA: deep drug-target binding affinity prediction. Bioinformatics 34, I821–I829 (2018)
Wang, S., et al.: MCN-CPI: multiscale convolutional network for compound-protein interaction prediction. Biomolecules 11(8), 1119 (2021)
Tsubaki, M., Tomii, K., Sese, J.: Compound-protein interaction prediction with end-to-end learning of neural networks for graphs and sequences. Bioinformatics 35(2), 309–318 (2019)
Li, S., Wan, F., Shu, H., Jiang, T., Zhao, D., Zeng, J.: MONN: a multi-objective neural network for predicting compound-protein interactions and affinities. Cell Syst. 10(4), 308–322 (2020)
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems 30 (2017)
Chen, L., et al.: TransformerCPI: improving compound-protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics 36(16), 4406–4414 (2020)
Chen, W., Chen, G., Zhao, L., Chen, C.Y.C.: Predicting drug-target interactions with deep-embedding learning of graphs and sequences. J. Phys. Chem. A 125(25), 5633–5642 (2021)
Wen, J., Gan, H., Yang, Z., Zhou, R., Zhao, J., Ye, Z.: Mutual-DTI: a mutual interaction feature-based neural network for drug-target protein interaction prediction. Math. Biosci. Eng. 20(6), 10610–10625 (2023)
Qian, Y., Li, X., Wu, J., Zhou, A., Xu, Z., Zhang, Q.: Picture-word order compound protein interaction: predicting compound-protein interaction using structural images of compounds. J. Comput. Chem. 43(4), 255–264 (2022)
Qian, Y., Wu, J., Zhang, Q.: CAT-CPI: combining CNN and transformer to learn compound image features for predicting compound-protein interactions. Front. Mol. Biosci. 9, 963912 (2022)
Costa, F., De Grave, K.: Fast neighborhood subgraph pairwise distance kernel. In: Proceedings of the 26th International Conference on Machine Learning, pp. 255–262. Omnipress, Madison, WI, USA (2010)
Dong, Q.W., Wang, X., Lin, L.: Application of latent semantic analysis to protein remote homology detection. Bioinformatics 22(3), 285–290 (2006)
Dauphin, Y.N., Fan, A., Auli, M., Grangier, D.: Language modeling with gated convolutional networks. In: Proceedings of the 34th International Conference on Machine Learning, pp. 933–941. PMLR, Sydney, NSW, Australia (2017)
Davis, M.I., et al.: Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29(11), 1046–1051 (2011)
Wishart, D., et al.: DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672 (2006)
Zhao, Q., Zhao, H., Zheng, K., Wang, J.: HyperAttentionDTI: improving drug-protein interaction prediction by sequence-based deep learning with attention mechanism. Bioinformatics 38(3), 655–662 (2022)
Acknowledgements
This work is supported by the High-level Talents Fund of Hubei University of Technology under grant No. GCRC2020016, Open Research Fund Program of State Key Laboratory of Biocatalysis and Enzyme Engineering under grant No. SKLBEE2021020 and SKLBEE2020020.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Wen, J., Gan, H., Yang, Z., Shi, M., Wang, J. (2024). MView-DTI: A Multi-view Feature Fusion-Based Approach for Drug-Target Protein Interaction Prediction. In: Luo, B., Cheng, L., Wu, ZG., Li, H., Li, C. (eds) Neural Information Processing. ICONIP 2023. Communications in Computer and Information Science, vol 1964. Springer, Singapore. https://doi.org/10.1007/978-981-99-8141-0_30
Download citation
DOI: https://doi.org/10.1007/978-981-99-8141-0_30
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-8140-3
Online ISBN: 978-981-99-8141-0
eBook Packages: Computer ScienceComputer Science (R0)