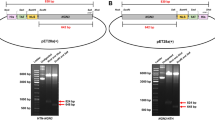
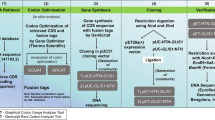
Abstract
NANOG is important for the early stages of embryogenesis and embryonic stem cell maintenance. It is also involved in the nuclear reprogramming necessary for forming induced pluripotent stem cells, thus offering enormous potential for cell therapies. Here, the expression and purification of human NANOG protein from the soluble fraction of bacterial cell lysate are reported. To produce fusion tagged transducible version of NANOG protein, we fused the human NANOG cDNA sequence to different fusion tags for affinity purification and efficient cell and nuclear delivery. Subsequently, the optimal expression conditions were identified, and the recombinant fusion protein was purified to homogeneity with a retained secondary structure. Additionally, NANOG is linked to cellular multipotency, which plays crucial roles in stemness, tumor progression and migration, thus highlighting its potential role as a therapeutic target for a myriad of malignancies such as cervical cancer. Therefore, we investigated the effect of application of the HTN-NANOG fusion protein on HeLa cells. Upon protein transduction in HeLa cells, the cell migration rate was augmented in the presence of NANOG. Furthermore, we observed the downregulation of p27 in HeLa cells in the presence of NANOG at mRNA and protein levels. From a future perspective, this bioactive form of human NANOG protein can be used to understand the molecular roles of NANOG in several cellular processes associated with cancer and induced pluripotent stem cells.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Booth HAF, Holland PWH (2004) Eleven daughters of NANOG. Genomics 84(2):229–238. https://doi.org/10.1016/j.ygeno.2004.02.014
Theunissen TW, Costa Y, Radzisheuskaya A, van Oosten AL, Lavial F, Pain B et al (2011) Reprogramming capacity of Nanog is functionally conserved in vertebrates and resides in a unique homeodomain. Development 138(22):4853–4865. https://doi.org/10.1242/dev.068775
Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113(5):643–655. https://doi.org/10.1016/s0092-8674(03)00392-1
Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K et al (2003) The homeoprotein nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113(5):631–642. https://doi.org/10.1016/S0092-8674(03)00393-3
Komatsu K, Fujimori T (2015) Multiple phases in regulation of Nanog expression during pre-implantation development. Dev Growth Differ 57(9):648–656. https://doi.org/10.1111/dgd.12244
Chambers I, Silva J, Colby D, Nichol J, Nijmeijer B, Robertson M et al (2007) Nanog safeguards pluripotency and mediates germline development. Nature 450(7173):1230–1234. https://doi.org/10.1038/nature06403
Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T (2005) Nanog expression in mouse germ cell development. Gene Expr Patterns 5(5):639–646. https://doi.org/10.1016/j.modgep.2005.03.001
Lavial F, Acloque H, Bertocchini F, MacLeod DJ, Boast S, Bachelard E et al (2007) The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development 134(19):3549–3563. https://doi.org/10.1242/dev.006569
Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O et al (2009) Nanog is the gateway to the pluripotent ground state. Cell 138(4):722–737. https://doi.org/10.1016/j.cell.2009.07.039
Messerschmidt DM, Kemler R (2010) Nanog is required for primitive endoderm formation through a non-cell autonomous mechanism. Dev Biol 344(1):129–137. https://doi.org/10.1016/j.ydbio.2010.04.020
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP et al (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122(6):947–956. https://doi.org/10.1016/j.cell.2005.08.020
Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X et al (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38(4):431–440. https://doi.org/10.1038/ng1760
Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444(7117):364–368. https://doi.org/10.1038/nature05284
Silva J, Chambers I, Pollard S, Smith A (2006) Nanog promotes transfer of pluripotency after cell fusion. Nature 441(7096):997–1001. https://doi.org/10.1038/nature04914
Hanna J, Saha K, Pando B, Van Zon J, Lengner CJ, Creyghton MP et al (2009) Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462(7273):595–601. https://doi.org/10.1038/nature08592
Gingold JA, Fidalgo M, Guallar D, Lau Z, Sun Z, Zhou H et al (2014) A genome-wide RNAi screen identifies opposing functions of snai1 and snai2 on the nanog dependency in reprogramming. Mol Cell 56(1):140–152. https://doi.org/10.1016/j.molcel.2014.08.014
Theunissen TW, Van Oosten AL, Castelo-Branco G, Hall J, Smith A, Silva JCR (2011) Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr Biol 21(1):65–71. https://doi.org/10.1016/j.cub.2010.11.074
Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang Wet al (2008) Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell 3(5):475–479. https://doi.org/10.1016/j.stem.2008.10.002
Schwarz BA, Bar-Nur O, Silva JCR, Hochedlinger K (2014) Nanog is dispensable for the generation of induced pluripotent stem cells. Curr Biol 24(3):347–350. https://doi.org/10.1016/j.cub.2013.12.050
Carter AC, Davis-Dusenbery BN, Koszka K, Ichida JK, Eggan K (2014) Nanog-independent reprogramming to iPSCs with canonical factors. Stem Cell Rep 2(2):119–126. https://doi.org/10.1016/j.stemcr.2013.12.010
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S et al (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318(5858):1917–1920. https://doi.org/10.1126/science.1151526
Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K (2008) A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3(3):340–345. https://doi.org/10.1016/j.stem.2008.08.003
Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C et al (2009) Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell 5(4):434–441. https://doi.org/10.1016/j.stem.2009.08.021
Li Y, Zhao H, Lan F, Lee A, Chen L, Lin C et al (2010) Generation of human-induced pluripotent stem cells from gut mesentery-derived cells by ectopic expression of OCT4/SOX2/NANOG. Cell Reprogramming (Formerly” Cloning and Stem Cells”), 12(3):237–247. https://doi.org/10.1089/cell.2009.0103
Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R et al (2008) Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci 105(8):2883–2888. https://doi.org/10.1073/pnas.0711983105
Moon J-H, Yun W, Kim J, Hyeon S, Kang PJ, Park G et al (2013) Reprogramming of mouse fibroblasts into induced pluripotent stem cells with Nanog. Biochem Biophys Res Commun 431(3):444–449. https://doi.org/10.1016/j.bbrc.2012.12.149
Borgohain MP, Haridhasapavalan KK, Dey C, Adhikari P, Thummer RP (2019) An insight into DNA-free reprogramming approaches to generate integration-free induced pluripotent stem cells for prospective biomedical applications. Stem Cell Rev Rep 15(2):286–313. https://doi.org/10.1007/s12015-018-9861-6
Haridhasapavalan KK, Borgohain MP, Dey C, Saha B, Narayan G, Kumar S, Thummer RP (2019) An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene 686:146–159. https://doi.org/10.1016/j.gene.2018.11.069
Dey C, Raina K, Haridhasapavalan KK, Thool M, Sundaravadivelu PK, Adhikari P et al (2021) An overview of reprogramming approaches to derive integration-free induced pluripotent stem cells for prospective biomedical applications. Recent Adv iPSC Technol 231–287. https://doi.org/10.1016/B978-0-12-822231-7.00011-4
Fischedick G, Wu G, Adachi K, Araúzo-Bravo MJ, Greber B, Radstaak M et al (2014) Nanog induces hyperplasia without initiating tumors. Stem Cell Res 13(2):300–315. https://doi.org/10.1016/j.scr.2014.08.001
Ezeh UI, Turek PJ, Reijo RA, Clark AT (2005) Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer: Interdiscip Int J Am Cancer Soc 104(10):2255–2265. https://doi.org/10.1002/cncr.21432
Bussolati B, Bruno S, Grange C, Ferrando U, Camussi G (2008) Identification of a tumor-initiating stem cell population in human renal carcinomas. FASEB J 22(10):3696–3705. https://doi.org/10.1096/fj.08-102590
Ye F, Zhou C, Cheng Q, Shen J, Chen H (2008) Stem-cell-abundant proteins Nanog, Nucleostemin and Musashi1 are highly expressed in malignant cervical epithelial cells. BMC Cancer 8(1):1–5. https://doi.org/10.1186/1471-2407-8-108
Thool M, Dey C, Bhattacharyya S, Sudhagar S, Thummer RP (2021) Generation of a recombinant stem cell-specific human SOX2 protein from Escherichia coli under native conditions. Mol Biotechnol 63(4):327–338. https://doi.org/10.1007/s12033-021-00305-y
Haridhasapavalan KK, Sundaravadivelu PK, Bhattacharyya S, Ranjan SH, Raina K, Thummer RP (2021) Generation of cell-permeant recombinant human transcription factor GATA4 from E. coli. Bioprocess Biosyst Eng 44(6):1131–1146. https://doi.org/10.1007/s00449-021-02516-8
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254. https://doi.org/10.1006/abio.1976.9999
Micsonai A, Wien F, Bulyáki É, Kun J, Moussong É, Lee Y-H et al (2018) BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res 46(W1):W315–W322. https://doi.org/10.1093/nar/gky497
Micsonai A, Bulyáki É, Kardos J (2021) BeStSel: from secondary structure analysis to protein fold prediction by circular dichroism spectroscopy. In: Structural genomics. Springer, Berlin, pp 175–189. https://doi.org/10.1007/978-1-0716-0892-0_11
Dey C, Thool M, Bhattacharyya S, Sudhagar S, Thummer RP (2021) Generation of biologically active recombinant human OCT4 protein from E. coli. 3 Biotech 11(5):1–16. https://doi.org/10.1007/s13205-021-02758-z
Narayan G, Agrawal A, Joshi N, Gogoi R, Nagotu S, Thummer RP (2021) Protein production and purification of a codon-optimized human NGN3 transcription factor from E. coli. Protein J 40(6):891–906. https://doi.org/10.1007/s10930-021-10020-x
Patel TB, Bertics PJ (2006) Epidermal growth factor: methods and protocols, vol 327. Springer Science & Business Media
Dey C, Venkatesan V, Thummer RP (2022) Identification of optimal expression parameters and purification of a codon-optimized human GLIS1 transcription factor from Escherichia coli. Mol Biotechnol 64(1):42–56. https://doi.org/10.1007/s12033-021-00390-z
Grada A, Otero-Vinas M, Prieto-Castrillo F, Obagi Z, Falanga V (2017) Research techniques made simple: analysis of collective cell migration using the wound healing assay. J Investig Dermatology 137(2):e11–e16. https://doi.org/10.1016/j.jid.2016.11.020
Haridhasapavalan KK, Das NJ, Thummer RP (2022) Generation of a transducible version of a bioactive recombinant human TBX5 transcription factor from E. Coli. Curr Res Biotechnol 4:66–77
Jauch R, Ng CKL, Saikatendu KS, Stevens RC, Kolatkar PR (2008) Crystal structure and DNA binding of the homeodomain of the stem cell transcription factor Nanog. J Mol Biol 376(3):758–770. https://doi.org/10.1016/j.jmb.2007.11.091
Hu PF, Guan WJ, Li XC, Ma YH (2012) Construction of recombinant proteins for reprogramming of endangered Luxi cattle fibroblast cells. Mol Biol Rep 39(6):7175–7182. https://doi.org/10.1007/s11033-012-1549-4
Yu M, Lian S, Han H, Yu K, Li G, Lian Z, Li N (2013) Four recombinant pluripotency transcriptional factors containing a protein transduction domain maintained the in vitro pluripotency of chicken embryonic stem cells. Sci China Life Sci 56(1):40–50. https://doi.org/10.1007/s11427-012-4426-4
Zhang H, Ma Y, Gu J, Liao B, Li J, Wong J, Jin Y (2012) Reprogramming of somatic cells via TAT-mediated protein transduction of recombinant factors. Biomaterials 33(20):5047–5055. https://doi.org/10.1016/j.biomaterials.2012.03.061
Yang WC, Patel KG, Lee J, Ghebremariam YT, Wong HE, Cooke JP, Swartz JR (2009) Cell-free production of transducible transcription factors for nuclear reprogramming. Biotechnol Bioeng 104(6):1047–1058. https://doi.org/10.1002/bit.22517
Yang WC, Welsh JP, Lee J, Cooke JP, Swartz JR (2011) Solubility partner IF2 Domain I enables high yield synthesis of transducible transcription factors in Escherichia coli. Protein Expr Purif 80(1):145–151. https://doi.org/10.1016/j.pep.2011.06.017
Ha SC, Pereira JH, Jeong JH, Huh JH, Kim SH (2009) Purification of human transcription factors Nanog and Sox2, each in complex with Skp, an Escherichia coli periplasmic chaperone. Protein Expr Purif 67(2):164–168. https://doi.org/10.1016/j.pep.2009.05.003
Kelly SM, Jess TJ, Price NC (2005) How to study proteins by circular dichroism. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1751(2):119–139. https://doi.org/10.1016/j.bbapap.2005.06.005
Micsonai A, Wien F, Kernya L, Lee Y-H, Goto Y, Réfrégiers M, Kardos J (2015) Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci 112(24):E3095–E3103. https://doi.org/10.1073/pnas.1500851112
Yu Z, Huang Z, Lung ML (2013) Subcellular fractionation of cultured human cell lines. Bio-Protoc 3(9):e754–e754
Do HJ, Lim HY, Kim JH, Song H, Chung HM, Kim JH (2007) An intact homeobox domain is required for complete nuclear localization of human Nanog. Biochem Biophys Res Commun 353(3):770–775. https://doi.org/10.1016/j.bbrc.2006.12.100
Chang DF, Tsai SC, Wang XC, Xia P, Senadheera D, Lutzko C (2009) Molecular characterization of the human NANOG protein. Stem Cells 27(4):812–821. https://doi.org/10.1634/stemcells.2008-0657
Zhang J, Wang X, Chen B, Suo G, Zhao Y, Duan Z, Dai J (2005) Expression of Nanog gene promotes NIH3T3 cell proliferation. Biochem Biophys Res Commun 338(2):1098–1102. https://doi.org/10.1016/j.bbrc.2005.10.071
Peitz M, Münst B, Thummer RP, Helfen M, Edenhofer F (2014) Cell-permeant recombinant Nanog protein promotes pluripotency by inhibiting endodermal specification. Stem cell Res 12(3):680–689. https://doi.org/10.1016/j.scr.2014.02.006
Gu TT, Liu SY, Zheng PS (2012) Cytoplasmic NANOG-positive stromal cells promote human cervical cancer progression. Am J Pathol 181(2):652–661. https://doi.org/10.1016/j.ajpath.2012.04.008
Ding Y, Yu AQ, Li CL, Fang J, Zeng Y, Li DS (2014) TALEN-mediated Nanog disruption results in less invasiveness, more chemosensitivity and reversal of EMT in Hela cells. Oncotarget 5(18):8393–8401. https://doi.org/10.18632/oncotarget.2298
Ding Y, Yu AQ, Wang XL, Guo XR, Yuan YH, Li DS (2016) Forced expression of Nanog with mRNA synthesized in vitro to evaluate the malignancy of HeLa cells through acquiring cancer stem cell phenotypes. Oncol Rep 35(5):2643–2650. https://doi.org/10.3892/or.2016.4639
Münst B, Thier MC, Winnemöller D, Helfen M, Thummer RP, Edenhofer F (2016) Nanog induces suppression of senescence through downregulation of p27KIP1 expression. J Cell Sci 129(5):912–920. https://doi.org/10.1242/jcs.167932
Oh JH, Do HJ, Yang HM, Moon SY, Cha KY, Chung HM, Kim JH (2005) Identification of a putative transactivation domain in human Nanog. Exp Mol Med 37(3):250–254. https://doi.org/10.1038/emm.2005.33
Jeter CR, Yang T, Wang J, Chao HP, Tang DG (2015) Concise review: NANOG in cancer stem cells and tumor development: an update and outstanding questions. Stem Cells 33(8):2381–2390. https://doi.org/10.1002/stem.2007
Patra SK, Vemulawada C, Soren MM, Sundaray JK, Panda MK, Barman HK (2018) Molecular characterization and expression patterns of Nanog gene validating its involvement in the embryonic development and maintenance of spermatogonial stem cells of farmed carp, Labeo rohita. J Anim Sci Biotechnol 9(1):1–17. https://doi.org/10.1186/s40104-018-0260-2
Stuart HT, Van Oosten AL, Radzisheuskaya A, Martello G, Miller A, Dietmann S et al (2014) NANOG amplifies STAT3 activation and they synergistically induce the naive pluripotent program. Curr Biol 24(3):340–346. https://doi.org/10.1016/j.cub.2013.12.040
Choi KJ, Quan MD, Qi C, Lee JH, Tsoi PS, Zahabiyon M et al (2022) NANOG prion-like assembly mediates DNA bridging to facilitate chromatin reorganization and activation of pluripotency. Nature Cell Biol 24(5):737–747. https://doi.org/10.1038/s41556-022-00896-x
Bothmann H, Plückthun A (1998) Selection for a periplasmic factor improving phage display and functional periplasmic expression. Nat Biotechnol 16(4):376–380. https://doi.org/10.1038/nbt0498-376
Hayashi Y, Caboni L, Das D, Yumoto F, Clayton T, Deller MC et al (2015) Structure-based discovery of NANOG variant with enhanced properties to promote self-renewal and reprogramming of pluripotent stem cells. Proc Natl Acad Sci U S A 112(15):4666–4671. https://doi.org/10.1073/pnas.1502855112
Bosnali M, Edenhofer F (2008) Generation of transducible versions of transcription factors Oct4 and Sox2. Biol Chem 389(7):851–861. https://doi.org/10.1515/BC.2008.106
Stock K, Nolden L, Edenhofer F, Quandel T, Brüstle O (2010) Transcription factor-based modulation of neural stem cell differentiation using direct protein transduction. Cell Mol Life Sci 67(14):2439–2449. https://doi.org/10.1007/s00018-010-0347-1
Thier M, Münst B, Edenhofer F (2011) Exploring refined conditions for reprogramming cells by recombinant Oct4 protein. Int J Dev Biol 54(11–12):1713–1721. https://doi.org/10.1387/ijdb.103193mt
Thier M, Münst B, Mielke S, Edenhofer F (2012) Cellular reprogramming employing recombinant Sox2 protein. Stem Cells Int. https://doi.org/10.1155/2012/549846
Narayan G, Sundaravadivelu PK, Agrawal A, Gogoi R, Nagotu S, Thummer RP (2021) Soluble expression, purification, and secondary structure determination of human PDX1 transcription factor. Protein Expr Purif 180:105807. https://doi.org/10.1016/j.pep.2020.105807
Haridhasapavalan KK, Sundaravadivelu PK, Voorkara U, Kaveeshwar V, Thummer RP (2022) Generation of the recombinant version of a bioactive human MEF2C transcription factor from E. coli. In: Healthcare research and related technologies-proceedings of NERC. Springer Nature
Haridhasapavalan KK, Sundaravadivelu PK, Thummer RP (2020) Codon optimization, cloning, expression, purification, and secondary structure determination of human ETS2 transcription factor. Mol Biotechnol 62(10):485–494. https://doi.org/10.1007/s12033-020-00266-8
Haridhasapavalan KK, Ranjan SH, Bhattacharyya S, Thummer RP (2021) Soluble expression, purification, and secondary structure determination of human MESP1 transcription factor. Appl Microbiol Biotechnol 105(6):2363–2376. https://doi.org/10.1007/s00253-021-11194-1
Haridhasapavalan KK, Sundaravadivelu PK, Joshi N, Das NJ, Mohapatra A, Voorkara U et al (2022) Generation of a recombinant version of a biologically active cell-permeant human HAND2 transcription factor from E. coli. Sci Rep 12(1):16129. https://doi.org/10.1038/s41598-022-19745-w
Acknowledgements
North Eastern Region—Biotechnology Programme Management Cell (BT/PR16655/NER/95/132/2015), DBT, Govt. of India funded this study.
Author information
Authors and Affiliations
Contributions
MT designed and performed the experiments, assembled and analyzed the data, wrote the manuscript; SS analyzed and interpreted the data; and RPT conceptualized and designed the study, analyzed the data, supervised the experiments, wrote the manuscript, and provided financial support. The final draft of the manuscript was approved by all the authors for publication.
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
None.
Conclusion
The purification of HTN-NANOG fusion protein from E. coli with a retained secondary structure and its bioactivity in human cells is demonstrated in this study. This study reported that human NANOG fusion protein enhanced proliferation, migration and downregulated p27 gene expression in HeLa cells, demonstrating its bioactivity. This biologically active human NANOG protein can be utilized to elucidate the biological function of NANOG in various cellular processes associated with cancer and induced pluripotent stem cells.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Thool, M., Sudhagar, S., Thummer, R.P. (2023). Soluble Expression and Purification of Biologically Active Human NANOG from Escherichia coli. In: Pandey, L.M., Gupta, R., Thummer, R.P., Kar, R.K. (eds) Healthcare Research and Related Technologies. NERC 2022. Springer, Singapore. https://doi.org/10.1007/978-981-99-4056-1_6
Download citation
DOI: https://doi.org/10.1007/978-981-99-4056-1_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-4055-4
Online ISBN: 978-981-99-4056-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)