Abstract
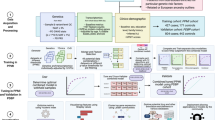
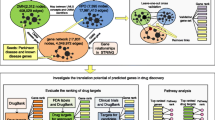
Weutilized interpretable deep learning methodologies to discern critical genes and latent biomarkers associated with Parkinson’s disease (PD). Gene expression data were collected from the GEO dataset, subjected to rigorous differential expression analysis to curate genes for subsequent scrutiny. Based on the P-Net and PASNet models, we have developed a pathway-related deep learning model that integrates PD-associated gene expression data with established biological pathways. This method has yielded satisfactory results, manifesting an Area Under the Curve (AUC) of 0.73 and an F1 score of 0.71, thereby efficaciously discriminating PD patients and bestowing novel insights into the pertinent biological pathways. Through interpretable deep learning models, we have identified potential biomarkers (XK, PDK1, TUBA4B, TP53) and their associated biological pathways (innate immune system, hemostasis, G protein-coupled receptor signaling pathway) related to Parkinson’s disease. The importance of these genes has been validated through external datasets and UPDRS III scores. Of particular significance is the XK gene, also known as Kell blood group precursor, and numerous XK gene mutations have been linked to the McLeod syndrome which exhibits symptomatic similarities with PD. Taken together, we identified several PD associated genes by explicable deep learning and bioinformatics methods, and XK gene was demonstrated a close correlation to PD.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
GBD 2016 Neurology Collaborators. Global, regional, and national burden of neurological disorders, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 18, 459–480 (2019)
Dorsey, E.R., Sherer, T., Okun, M.S., Bloem, B.R.: The emerging evidence of the Parkinson pandemic. J. Parkinsons Dis. 8, S3–S8 (2018)
Tysnes, O.-B., Storstein, A.: Epidemiology of Parkinson’s disease. J. Neural Transm. 124, 901–905 (2017)
Clarke, C.E.: Parkinson’s disease. BMJ 335, 441–445 (2007)
Rauschert, S., Raubenheimer, K., Melton, P.E., Huang, R.C.: Machine learning and clinical epigenetics: a review of challenges for diagnosis and classification. Clin. Epigenetics 12, 51 (2020)
Zhou, X., Chen, Y., Ip, F.C.F., Jiang, Y., Cao, H., Lv, G., et al.: Deep learning-based polygenic risk analysis for Alzheimer’s disease prediction. Commun. Med. 3, 49 (2023)
Shamir, R., Klein, C., Amar, D., Vollstedt, E.-J., Bonin, M., Usenovic, M., et al.: Analysis of blood-based gene expression in idiopathic Parkinson disease. Neurology 89, 1676–1683 (2017)
Locascio, J.J., Eberly, S., Liao, Z., Liu, G., Hoesing, A.N., Duong, K., et al.: Association between α-synuclein blood transcripts and early, neuroimaging-supported Parkinson’s disease. Brain 138, 2659–2671 (2015)
Scherzer, C.R., Eklund, A.C., Morse, L.J., Liao, Z., Locascio, J.J., Fefer, D., et al.: Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc. Natl. Acad. Sci. U.S.A. 104, 955–960 (2007)
Hao, J., Kim, Y., Kim, T.-K., Kang, M.: PASNet: pathway-associated sparse deep neural network for prognosis prediction from high-throughput data. BMC Bioinform. 19, 510 (2018)
Elmarakeby, H.A., Hwang, J., Arafeh, R., Crowdis, J., Gang, S., Liu, D., et al.: Biologically informed deep neural network for prostate cancer discovery. Nature 598, 348–352 (2021)
Ringnér, M.: What is principal component analysis? Nat. Biotechnol. 26, 303–304 (2008)
Ritchie, M.E., Phipson, B., Wu, D., Hu, Y., Law, C.W., Shi, W., et al.: Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015)
Jassal, B., Matthews, L., Viteri, G., Gong, C., Lorente, P., Fabregat, A., et al.: The reactome pathway knowledgebase. Nucleic Acids Res. 48, D498-503 (2020)
Vickers, A.J., Holland, F.: Decision curve analysis to evaluate the clinical benefit of prediction models. Spine J. 21, 1643–1648 (2021)
Farkas, S., et al.: Signal transduction pathway activity compensates dopamine D2/D3 receptor density changes in Parkinson’s disease: a preliminary comparative human brain receptor autoradiography study with [3H]raclopride and [35S]GTPγS. Brain Res. 9(1453), 56–63 (2012)
Tansey, M.G., Wallings, R.L., Houser, M.C., Herrick, M.K., Keating, C.E., Joers, V.: Inflammation and immune dysfunction in Parkinson disease. Nat. Rev. Immunol. 22(11), 657–673 (2022)
Bastide, M.F., Bido, S., Duteil, N., Bézard, E.: Striatal NELF-mediated RNA polymerase II stalling controls l-dopa induced dyskinesia. Neurobiol. Dis. 85, 93–98 (2016)
Harms, A.S., Ferreira, S.A., Romero-Ramos, M.: Periphery and brain, innate and adaptive immunity in Parkinson’s disease. Acta Neuropathol. 141, 527–545 (2021)
Schlachetzki, J.C., Winkler, J.: The innate immune system in Parkinson’s disease: a novel target promoting endogenous neuroregeneration. Neural Regen. Res. 10(5), 704–706 (2015)
Sulzer, D., et al.: T cells from patients with Parkinson’s disease recognize α-synuclein peptides. Nature 546(7660), 656–661 (2017)
Takeuchi, A., et al.: CRTAM determines the CD4+ cytotoxic T lymphocyte lineage. J. Exp. Med. 213(1), 123–138 (2016)
Earls, R.H., Lee, J.K.: The role of natural killer cells in Parkinson’s disease. Exp. Mol. Med. 52(9), 1517–1525 (2020)
Saulquin, X., Gastinel, L.N., Vivier, E.: Crystal structure of the human natural killer cell activating receptor KIR2DS2 (CD158j). J. Exp. Med. 197(7), 933–938 (2003)
Vallée, A., Lecarpentier, Y., Guillevin, R., Vallée, J.-N.: Thermodynamics in neurodegenerative diseases: interplay between canonical WNT/Beta-catenin pathway-PPAR gamma, energy metabolism and circadian rhythms. Neuromolecular Med. 20, 174–204 (2018)
Lu, T., Kim, P., Luo, Y.: Tp53 gene mediates distinct dopaminergic neuronal damage in different dopaminergic neurotoxicant models. Neural Regen. Res. 12, 1413–1417 (2017)
He, Z.-Q., Huan, P.-F., Wang, L., He, J.-C.: Paeoniflorin ameliorates cognitive impairment in Parkinson’s disease via JNK/p53 signaling. Metab. Brain Dis. 37, 1–14 (2022). https://doi.org/10.1007/s11011-022-00937-2
Daniels, G.L., Weinauer, F., Stone, C., Ho, M., Green, C.A., Jahn-Jochem, H., et al.: A combination of the effects of rare genotypes at the XK and KEL blood group loci results in absence of Kell system antigens from the red blood cells. Blood 88, 4045–4050 (1996)
Acknowledgment
We would like to express our heartfelt gratitude to the National Natural Science Foundation of China for providing the financial support through grant No. 32060157 and 82360098, Central Guiding Local Science and Technology Development Fund Projects (ZY20230103), Graduate research project of Guilin Medical University No. GYYK2022020, which made this study possible.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Zhang, Y. et al. (2024). Identification of Parkinson’s Disease Associated Genes Through Explicable Deep Learning and Bioinformatic. In: Huang, DS., Premaratne, P., Yuan, C. (eds) Applied Intelligence. ICAI 2023. Communications in Computer and Information Science, vol 2014. Springer, Singapore. https://doi.org/10.1007/978-981-97-0903-8_14
Download citation
DOI: https://doi.org/10.1007/978-981-97-0903-8_14
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-97-0902-1
Online ISBN: 978-981-97-0903-8
eBook Packages: Computer ScienceComputer Science (R0)