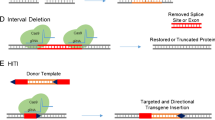
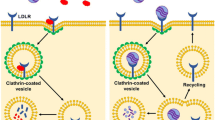
Abstract
Fatty liver disease is characterized as nonalcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD). Fatty liver disease is one of the most common causes of chronic liver disease worldwide among adults and children. It is characterized by excessive fat accumulation in the liver cells. It has a genetically heterogenous background with complex pathogenesis and progressions and is accompanied by significant morbidity, mortality, and healthcare costs. NAFLD’s risk factors include metabolic syndrome, abdominal obesity, type 2 diabetes, and atherogenic dyslipidemia. ALD is associated with the excessive consumption of alcohol. Here, we describe the functions of various proteins encoded by gene variants contributing to the pathogenesis of nonalcoholic fatty liver disease and alcoholic fatty liver disease. Advancements in genome engineering technology have generated various in vivo and in vitro fatty liver disease models reflecting the genetic abnormalities contributing toward fatty liver disease. We will discuss currently developed different ALD and NAFLD models using the clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) genome editing tool.
Furthermore, we will also discuss the salient features of CRISPR/Cas9 editing technology and Cas9 variants such as prime and base editors to replicate genetic topographies linked specifically to ALD and NAFLD. The advantages and limitations of currently available genome delivery methods necessary for optimal gene editing will also be discussed in this review. This review will provide the essential guidance for appropriate genome editing tool selection and proper gene delivery approaches for the effective development of ALD and NAFLD models, leading to the development of clinical therapeutics for fatty liver disease.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Brunt EM, Wong VW, Nobil V, Day CP, Sookoian S, Maher JJ, Bugianesi E, Sirlin CB, Neuschwander-Tetri BA, Rinella ME (2015) Nonalcoholic fatty liver disease. Nat Rev Dis Primers 2015:1–61
Anstee QM, Targher G, Day CP (1965) Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol 10:330–344
Lieber CS, Jones DP, Decarli LM (1965) Effects of prolonged ethanol intake: production of fatty liver despite adequate diets. J Clin Investig 44:1009–1021
Lieber CS, Decarli LM (1976) Animal models of ethanol dependence and liver injury in rats and baboons. Fed Proc 35:1232–1236
Brandon-Warner E, Schrum LW, Schmidt CM (2012) Rodent models of alcoholic liver disease: of mice and men. Alcohol 46(8):715–725
Bellentani S, Saccoccio G, Costa G (1997) Drinking habits as cofactors of risk for alcohol induced liver damage. Gut 41(6):845–850
Becker U, Deis A, Sorensen T (1996) Prediction of risk of liver disease by alcohol intake, sex, and age: a prospective population study. Hepatology 23(5):1025–1029
Hirschhorn JN, Gajdos ZK (2011) Genome-wide association studies: results from the first few years and potential implications for clinical medicine. Annu Rev Med 62:11–24
Sato N, Lindros KO, Baraona E (2001) Sex difference in alcohol-related organ injury. Alcohol Clin Exp Res 25(5):40S–45S
Stinson FS, Grant BF, Dufour MC (2001) The critical dimension of ethnicity in liver cirrhosis mortality statistics. Alcohol Clin Exp Res 25:1181–1187
Reed T, Page WF, Viken RJ (1996) Genetic predisposition to organ-specific endpoints of alcoholism. Alcohol Clin Exp Res 20(9):1528–1533
Hrubec Z, Omenn GS (1981) Evidence of genetic predisposition to alcoholic cirrhosis and psychosis: twin concordances for alcoholism and its biological end points by zygosity among male veterans. Alcohol Clin Exp Res 5(2):207–215
Whitfield JB, Rahman K, Haber PS (2015) Brief report: genetics of alcoholic cirrhosis-GenomALC multinational study. Alcohol Clin Exp Res 39:836–842
Said A, Williams J, Holden J (2004) The prevalence of alcohol-induced liver disease and hepatitis C and their interaction in a tertiary care setting. Clin Gastroenterol Hepatol 2(10):928–934
Anstee QM, Day CP (2013) The genetics of NAFLD. Nat Rev Gastroenterol Hepatol 10:645–655
Masuoka HC, Chalasani N (2013) Nonalcoholic fatty liver disease: an emerging threat to obese and diabetic individuals. Ann N Y Acad Sci 1281(1):106–122
Sanyal AJ, Brunt EM, Kleiner DE (2011) Endpoints and clinical trial design for nonalcoholic steatohepatitis. Hepatology 54(1):344–353
Bettermann K, Hohensee T, Haybaeck J (2014) Steatosis and steatohepatitis: complex disorders. Int J Mol Sci 15(6):9924–9944
Haybaeck J, Stumptner C, Thueringer A (2012) Genetic background effects of keratin 8 and 18 in a DDC-induced hepatotoxicity and Mallory-Denk body formation mouse model. Lab Investig 92:857–867
Zatloukal B, Kufferath I, Thueringer A (2014) Sensitivity and specificity of in situ proximity ligation for protein interaction analysis in a model of steatohepatitis with Mallory-Denk bodies. PLoS ONE 9(5):e96690
Younossi ZM, Koenig AB, Abdelatif D (2016) Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64(1):73–84
Bettermann K, Mehta AK, Hofer EM (2016) Keratin 18-deficiency results in steatohepatitis and liver tumors in old mice: A model of steatohepatitis-associated liver carcinogenesis. Oncotarget 7(45):73309–73322
Golob-Schwarzl N, Bettermann K, Mehta AK (2019) High keratin 8/18 ratio predicts aggressive hepatocellular cancer phenotype. Transl Oncol 12:256–268
Mikolasevic I, Milic S, Turk Wensveen T (2016) Nonalcoholic fatty liver disease - a multisystem disease? World J Gastroenterol 22(43):9488–9505
Severson TJ, Besur S, Bonkovsky HL (2016) Genetic factors that affect nonalcoholic fatty liver disease: a systematic clinical review. World J Gastroenterol 22(29):6742–6756
Younes R, Bugianesi E (2019) NASH in lean individuals. Semin Liver Dis 39(1):86–95
Kim HJ, Kim HJ, Lee KE (2004) Metabolic significance of nonalcoholic fatty liver disease in nonobese, nondiabetic adults. Arch Intern Med 164(19):2169–2175
Sookoian S, Pirola CJ (2017) Genetic predisposition in nonalcoholic fatty liver disease. Clin Mol Hepatol 23(1):1–12
Loomba R, Schork N, Chen CH (2015) Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology 149:1784–1793
Machado MV, Cortez-Pinto H (2014) Non-alcoholic fatty liver disease: what the clinician needs to know. World J Gastroenterol 20:12956–12980
Cardon LR, Bell JI (2001) Association study designs for complex diseases. Nat Rev Genet 2:91–99
Hirschhorn JN (2009) Genomewide association studies– illuminating biologic pathways. N Engl J Med 360:1699–1701
Chalasani N, Guo X, Loomba R (2010) Genome-wide association study identifies variants associated with histologic features of nonalcoholic fatty liver disease. Gastroenterology 139:1567–1576
Liu YL, Reeves HL, Burt AD, Tiniakos D, McPherson S, Leathart JB, Allison ME, Alexander GJ, Piguet AC (2014) TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun 5:4309
Mahdessian H, Taxiarchis A, Popov S, Silveira A (2014) TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A 111(24):8913–8918
Buch S, Stickel F, Trepo E (2015) A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet 47:1443–1448
Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD (2011) Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet 7:1001324
Petta S, Miele L, Bugianesi E, Camma C, Rosso C, Boccia S, Cabibi D, Di Marco V, Grimaudo S, Grieco A (2014) Glucokinase regulatory protein gene polymorphism affects liver fibrosis in non-alcoholic fatty liver disease. PLoS ONE 9:e87523
Falleti E, Cussigh A, Cmet S (2016) PNPLA3 rs738409 and TM6SF2 rs58542926 variants increase the risk of hepatocellular carcinoma in alcoholic cirrhosis. Dig Liver Dis 48:69–75
Hayashi S, Watanabe J, Kawajiri K (1991) Genetic polymorphisms in the 50 flanking regions change transcriptional regulation of the human cytochrome P450 IIE1 gene. J Biochem 10:559–565
Buch S, Stickel F, Trepo E, Way M, Herrmann A, Nischalke HD, Brosch M, Rosendahl J, Berg T, Ridinger M (2015) A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet 47:1443–1448
Mancina RM, Dongiovanni P, Petta S, Pingitore P, Meroni M, Rametta R, Boren J, Montalcini T, Pujia A, Wiklund O (2016) The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology 150:1219–1230.e6
Viitasalo A, Eloranta AM, Atalay M, Romeo S, Pihlajamaki J, Lakka TA (2016) Association of MBOAT7 gene variant with plasma ALT levels in children: the PANIC study. Pediatr Res 80(5):651–655
Donati B, Dongiovanni P, Romeo S, Meroni M, McCain M, Miele L, Petta S, Maier S, Rosso C, De Luca L (2017) MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci Rep 7:4492
Salley TN, Mishra M, Tiwari S, Jadhav A, Ndisang JF (2013) The heme oxygenase system rescues hepatic deterioration in the condition of obesity co-morbid with type-2 diabetes. PLoS ONE 8:e79270
Hinds TD Jr, Sodhi K, Meadows C, Fedorova L, Puri N, Kim DH, Peterson SJ, Shapiro J, Abraham NG, Kappas A (2014) Increased HO-1 levels ameliorate fatty liver development through a reduction of heme and recruitment of FGF21. Obesity 22:705–712
Tian C, Stokowski RP, Kershenobich D (2010) Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet 42:21–23
Salameh H, Raff E, Erwin A (2015) PNPLA3 gene polymorphism is associated with predisposition to and severity of alcoholic liver disease. Am J Gastroenterol 110:846–856
Kozlitina J, Smagris E, Stender S (2014) Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 46:352–356
Sookoian S, Castaño GO, Scian R, Mallardi P, Fernández Gianotti T, Burgueño AL, San Martino J, Pirola CJ (2015) Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology 61(2):515–525
Chen X, Zhou P, De L, Li B (2019) The roles of transmembrane 6 superfamily member 2 rs58542926 polymorphism in chronic liver disease: a meta-analysis of 24,147 subjects. Mol Genet Genom Med 7(8):e824
DiStefano JK, Kingsley C, Craig Wood G, Chu X, Argyropoulos G, Still CD, Doné SC, Legendre C, Tembe W, Gerhard GS (2015) Genome-wide analysis of hepatic lipid content in extreme obesity. Acta Diabetol 52(2):373–382
Way M, Atkinson S, McQuillin A (2015) A functional variant in tm6sf2 associates with alcohol related cirrhosis risk in a British and Irish population. J Hepatol 62(Suppl 2):S772
Pirola CJ, Flichman D, Dopazo H, Fernández Gianotti T, San Martino J, Rohr C, Garaycoechea M, Gazzi C, Castaño GO, Sookoian S (2018) A rare nonsense mutation in the glucokinase regulator gene is associated with a rapidly progressive clinical form of nonalcoholic steatohepatitis. Hepatol Commun 2(9):1030–1036
Li J, Zhao Y, Zhang H, Hua W, Jiao W, Du X, Rui J, Li S, Teng H, Shi B, Yang X, Zhu L (2020) Contribution of Rs780094 and Rs1260326 polymorphisms in GCKR gene to nonalcoholic fatty liver disease: a meta-analysis involving 26,552 participants. Endocr Metab Immune Disord 20:1696–1708
Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH (2008) Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 40:1461–1465
Liu YL, Patman GL, Leathart JB, Piguet AC, Burt AD, Dufour JF, Day CP, Daly AK, Reeves HL, Anstee QM (2014) Carriage of the PNPLA3 rs738409 C > G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol 61:75–81
Xu R, Tao A, Zhang S, Deng Y, Chen G (2015) Association between patatin-like phospholipase domain containing 3 gene (PNPLA3) polymorphisms and nonalcoholic fatty liver disease: HuGE review and meta-analysis. Sci Rep 5:9284
Chang PF, Lin YC, Liu K, Yeh SJ, Ni YH (2015) Heme oxygenase-1 gene promoter polymorphism and the risk of pediatric nonalcoholic fatty liver disease. Int J Obes 39:1236–1240
Gaj T, Gersbach CA, Barbas CF (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405
Alves-Bezerra M, Furey N, Johnson CG, Bissig KD (2019) Using CRISPR/Cas9 to model human liver disease. JHEP Rep 1:392–402
Barrangou R (2015) The roles of CRISPR-Cas systems in adaptive immunity and beyond. Curr Opin Immunol 32:36–41
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278
Shin HY, Wang C, Lee HK, Yoo KH, Zeng X, Kuhns T, Yang CM, Mohr T, Liu C, Hennighausen L (2017) CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat Commun 8:15464
Fan Y, Lu H, Guo Y, Zhu T, Garcia-Barrio MT, Jiang Z, Willer CJ, Zhang J, Chen YE (2016) Hepatic transmembrane 6 superfamily member 2 regulates cholesterol metabolism in mice. Gastroenterology 150:1208–1218
O’Hare EA, Yang R, Yerges-Armstrong LM, Sreenivasan U, McFarland R, Leitch CC, Wilson MH, Narina S, Gorden A, Ryan KA (2017) TM6SF2 rs58542926 impacts lipid processing in liver and small intestine. Hepatology 65:1526–1542
Luukkonen PK, Nick A, Holtta-Vuori M, Thiele C, Isokuortti E, Lallukka-Bruck S, Zhou Y, Hakkarainen A, Lundbom N, Peltonen M (2019) Human PNPLA3-I148M variant increases hepatic retention of polyunsaturated fatty acids. JCI Insight 4:e127902
Meroni M, Dongiovanni P, Longo M, Carli F, Baselli G, Rametta R, Pelusi S, Badiali S, Maggioni M, Gaggini M (2020) Mboat7 down-regulation by hyper-insulinemia induces fat accumulation in hepatocytes. EBioMedicine 52:102658
Trépo E, Romeo S, Zucman-Rossi J, Nahon P (2016) PNPLA3 gene in liver diseases. J Hepatol 65(2):399–412
Codner GF, Mianne J, Caulder A, Loeffler J, Fell R, King R, Allan AJ, Mackenzie M, Pike FJ, McCabe CV (2018) Application of long single-stranded DNA donors in genome editing: generation and validation of mouse mutants. Br Med J 16:70
Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D, Ma W (2018) Methodologies for improving HDR efficiency. Front Genet 9:691
Anzalone AV, Koblan LW, Liu DR (2020) Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38:824–844
Cox DB, Platt RJ, Zhang (2015) Therapeutic genome editing: prospects and challenges. Medicine 21(2):121–131
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017) Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551:464–471
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576:149–157
Mout R, Ray M, Yesilbag Tonga G, Lee YW, Tay T, Sasaki K, Rotello VM (2017) Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 11:2452–2458
Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S (2015) Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 208:44–53
Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154:1370–1379
Lau CH, Suh Y (2017) In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease. F1000Res 6:2153
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191
Wang Y, Wang M, Zheng T, Hou Y, Zhang P, Tang T, Wei J, Du Q (2020) Specificity profiling of CRISPR system reveals greatly enhanced off-target gene editing. Sci Rep 10:2269
Lindel F, Dodd CR, Weidner N, Noll M, Bergemann F, Behrendt R, Fischer S, Dietrich J, Cartellieri M, Hamann MV (2019) TraFo-CRISPR: enhanced genome engineering by transient foamy virus vector-mediated delivery of CRISPR/Cas9 components. Mol Ther Nucl Acids 18:708–726
Huo W, Zhao G, Yin J, Ouyang X, Wang Y, Yang C, Wang B, Dong P, Wang Z, Watari H (2017) Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer 8:57–64
Mansouri M, Ehsaei Z, Taylor V, Berger P (2017) Baculovirus-based genome editing in primary cells. Plasmid 90:5–9
Chen S, Lee B, Lee AY, Modzelewski AJ, He L (2016) Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J Biol Chem 291:14457–14467
Shin HY, Willi M, HyunYoo K, Zeng X, Wang C, Metser G, Hennighausen L (2016) Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat Genet 48:904–911
Kim S, Kim D, Cho SW, Kim J, Kim JS (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24:1012–1019
Horii T, Arai Y, Yamazaki M, Morita S, Kimura M, Itoh M, Abe Y, Hatada I (2014) Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci Rep 4:4513
Chuang CK, Chen CH, Huang CL, Su YH, Peng SH, Lin TY, Tai HC, Yang TS, Tu CF (2017) Generation of GGTA1 mutant pigs by direct pronuclear microinjection of CRISPR/Cas9 plasmid vectors. Anim Biotechnol 28:174–181
Crispo M, Mulet AP, Tesson L, Barrera N, Cuadro F, dos Santos-Neto PC, Nguyen TH, Creneguy A, Brusselle L, Anegon (2015) Efficient generation of myostatin knock-out sheep using CRISPR/Cas9 technology and microinjection into zygotes. PLoS ONE 10:e0136690
Lino CA, Harper JC, Carney JP, Timlin JA (2018) Delivering CRISPR: a review of the challenges and approaches. Drug Deliv 25:1234–1257
Li L, Hu S, Chen X (2018) Non-viral delivery systems for CRISPR/Cas9-based genome editing: challenges and opportunities. Biomaterials 171:207–218
Biagioni A, Laurenzana A, Margheri F, Chilla A, Fibbi G, Del Rosso M (2018) Delivery systems of CRISPR/Cas9-based cancer gene therapy. J Biol Eng 12:33
Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S (2017) In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun 8:14500
Yoo KH, Hennighausen L, Shin HY (2019) Dissecting tissue-specific super-enhancers by integrating genome-wide analyses and CRISPR/Cas9 genome editing. J Mam Gland Biol 24:47–59
Kim HK, Song M, Lee J, Menon AV, Jung S, Kang YM, Choi JW, Woo E, Koh HC, Nam JW (2017) In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat Methods 14:153–159
Ono C, Okamoto T, Abe T, Matsuura Y (2018) Baculovirus as a tool for gene delivery and gene therapy. Viruses 10:510
Volkman LE, Goldsmith PA (1983) In vitro survey of autographa californica nuclear polyhedrosis virus interaction with nontarget vertebrate host cells. Appl Environ Microbiol 45:1085–1093
Smagris E, BasuRay S, Huang LJ, Lai KM, Gromada J, Cohen JC, Hobbs HH (2015) Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 61:108–118
O’Hare EA, Yerges-Armstrong LM, Perry JA, Shuldiner AR, Zaghloul NA (2016) Assignment of functional relevance to genes at type 2 diabetes-associated loci through investigation of beta-cell mass deficits. Mol Endocrinol 30:429–445
Tanaka Y, Shimanaka Y, Caddeo A, Kubo T, Mao Y, Kubota T, Kubota N, Yamauchi T, Mancina RM, Baselli G (2020) LPIAT1/MBOAT7 depletion increases triglyceride synthesis fueled by high phosphatidylinositol turnover. Gut 70(1):180–193
Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH (2015) Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucl Acids 4:e264
Lee HK, Willi M, Miller SM, Kim S, Liu C, Liu DR, Hennighausen L (2018) Targeting fidelity of adenine and cytosine base editors in mouse embryos. Nat Commun 9:4804
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Hayat, U., Siddiqui, A.A., Farhan, M.L., Haris, A., Hameed, N. (2023). Genome Editing and Fatty Liver. In: Xiao, J. (eds) Genome Editing in Cardiovascular and Metabolic Diseases. Advances in Experimental Medicine and Biology, vol 1396. Springer, Singapore. https://doi.org/10.1007/978-981-19-5642-3_13
Download citation
DOI: https://doi.org/10.1007/978-981-19-5642-3_13
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-5641-6
Online ISBN: 978-981-19-5642-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)