Abstract
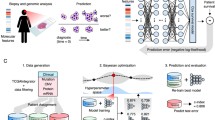
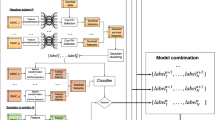
Deep learning (DL) is a leading paradigm in ML, which recently has brought huge improvements in benchmarks and provided principally new functionalities. The shift towards the deep extends the horizons in seemingly every field of clinical and bioinformatics analysis. Computational platform are exposed to a great volume of new methods promising improvements. Yet, there is a trade-off between the number of man/hours and the degree to which cutting edge advances in methodology are integrated into the routine procedure. Understanding why many of the new shiny methods published in the CS literature are not suitable to be applied in clinical research and making an explicit checklist would be of practical help. For example, when it comes to survival analysis for omics and clinicopathological variables, despite a rapidly growing number of architectures recently proposed, if one excludes image processing, the gain in efficiency and general benefits are somewhat unclear, recent reviews do not make a great emphasis on the deep paradigm either, and clinicians hardly ever use those. The consequences of these misunderstandings, which affects a number of published articles, results in the fact that the proposed methods are not attractive enough to enter applications. The example with the survival analysis motivates the need for computational platforms to work on the recommendations regarding (1) which methods should be considered as apt for a consideration to be integrated into the analysis practice for primary research articles, and (2) which literature reviews on cross-disciplinary topics are worth considering.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
McKenzie, T., et al.: Statement: an updated guideline for reporting systematic reviews. BMJ 372, 2021 (2020)
Antolini, L., et al.: A time-dependent discrimination index for survival data. Stat. Med. 24, 3927–3944 (2005)
Cabitza, A., Campagner, A., The need to separate the wheat from the chaff in medical informatics, Introducing a comprehensive checklist for the (self) assessment of medical AI studies. Int. J. Med. Inform. 153 (2021)
Ching, T., et al, Cox-nnet: an artificial neural network method for prognosis prediction of high-thoughput omics data. PloS Comput. Biol. 14(4) (2016)
Cox, D.R, Regression models and life-tables. J. R. Stat. Soc. Series B Methodol. 34, 187–220 (1972)
Deepa, P., Gunavathi, C.: A systematic review on machine learning and deep learning techniques in cancer survival prediction. Progress Biophys. Molecul. Biol. 174 (2022)
Sidorova, J., Lozano, J.J.: Appendix to “A systematic review on machine learning and deep learning, under review
techniques in cancer survival prediction”: Validation of Survival Methods
Faraggi, D., Simon, R.: A neural network for survival data. Stat. Med. 14(1), 72–73 (1995)
Grossman, R.L.: et al.: Toward a shared vision for cancer genomic data. N. Engl. J. Med. 375(12), 1109–1112 (2016)
Hao, et al.: Cox-PASNet: pathway-based sparse deep neural network for survival analysis, IEEE International Conference on Bioinformatics and Biomedicine, pp. 381–386, BIBM-2018
Heagerty, P.J., Zheng, Y.: Survival model predictive accuracy and ROC curves. Biometrics 61, 92–105 (2005)
Hu, F., et al.: A gene signature of survival prediction for kidney renal cell carcinoma by multi-omic data analysis. Int. J. Mol. Sci. 20(22), 5720 (2019)
Huang, et al.: SALMON: survival analysis learning with multi-omics neural networks on breast cancer. Front. Genet. (2019)
Sidorova, J., Lozano, J.J.: New and classical data processing: fruit and perils, poster. CIBER (2022)
Lee, S., Lim, H.: Review of statistical methods for survival analysis using genomic data. Genom. Inform. 17(4), e41 (2019)
Lee Ch., et al.: DeepHit: a deep learning approach to survival analysis with competing risks. In: Proceedings of 32nd AAAI Conference on Artificial Intelligence (2018)
Kim, S., et al.: Improved survival analysis by learning shared genomic information from pan-cancer data. Bioinformatics 36 (2020)
Kleinbaum, D.G., Klein, M.: Survival analysis, a self-learning text, 3rd edn. Springer, Statistics for Biology and Health (2012)
Kvamme, H., et al.: Time-to-event prediction with neural network and Cox regression. J. Mach. Learn. Res. 20 (2019)
Katzman, J., et al.: DeepSurv: personalised treatment recommender system using a Cox proportinal hazards deep neural network. BMC Med. Res. Methodol. 18, 24 (2018)
Sahu, et al.: Discovery of targets for immune-metabolic antitumor drugs identifies estrogen-related receptor alpha. Cancer Discov. 13(3), 672–701 (2023)
Salerno, S., Li, Y.: High-dimensional survival analysis: methods and applications. Ann. Rev. Statist. Appl. 10, 25–49 (2023)
Sidorova, J., Lozano, J.J.: New and classical data processing: fruit and perils, jornadas Cientificas, CIBER (2022)
Sidorova, J., Lozano, J.J.: A Survey of Survival Analysis with Deep Learning: Models, Applications and Challenges. under review
[SEER, http] https://seer.cancer.gov/causespecific/
Tong, L., et al.: Deep learning based feature-level integration pf multi-omics data for breast cancer patients survival analysis. BCM Med. Informa. Dec. Mak. 20, 225 (2020)
Yang, et al.: Identifying risk stratification associated with a cancer for overall survival by deep-learning based CoxPH. IEEE Access (7) (2019)
Yousefi, S., et al.: Predicting clinical outcomes from large scale cancer genomic profiles with deep survival models. Scientific Reports (2017)
Sidorova, J., et al.: Blood glucose estimation from voice: first review of successes and challenges. J. Voice (2020a)
Sidorova, J., et al.: Impact of diabetes mellitis on voice: a methodological commentary (2020b)
Hond, A., et al.: Perspectives of validation of clinical predictive algorithms, digital Medicine (2023)
Sidorova, J., Badia, T.: ESEDA: tool for enhanced emotion recognition and detection, Procs. AXMEDIS (2008)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Sidorova, J., Lozano, J.J. (2024). Need for Quality Auditing for Screening Computational Methods in Clinical Data Analysis, Including Revise PRISMA Protocols for Cross-Disciplinary Literature Reviews. In: Guarda, T., Portela, F., Diaz-Nafria, J.M. (eds) Advanced Research in Technologies, Information, Innovation and Sustainability. ARTIIS 2023. Communications in Computer and Information Science, vol 1935. Springer, Cham. https://doi.org/10.1007/978-3-031-48858-0_11
Download citation
DOI: https://doi.org/10.1007/978-3-031-48858-0_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-48857-3
Online ISBN: 978-3-031-48858-0
eBook Packages: Computer ScienceComputer Science (R0)