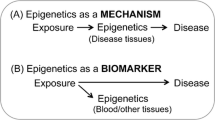
Abstract
In this chapter we consider the role of environmental factors on the epigenome. The importance of research into exposures that may alter epigenetic marks is now well recognized. Relations of exposures such as tobacco, alcohol, diet, endocrine disruptors, metals, and environmental contaminants with epigenetic states have been investigated and are reviewed here. We will briefly cover environmental exposures and imprinting and development, as well as discuss potential mechanisms for exposures to modify epigenetic states. Appropriate epidemiologic studies are crucial to understanding the true effect of environmental exposures on the human epigenome and this work is urgently needed to better understand the biology of epigenetic alterations which may constitute biology underlying risk for pathogenesis of disease. With a more comprehensive understanding of the effects of exposures on the epigenome (including consideration of genetic background), not only will the prediction of the toxic potential of new compounds be more readily achieved, but precision prevention and intervention strategies also may be developed.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- ADHD:
-
attention deficit hyperactivity disorder
- ART:
-
assisted reproductive technology
- AUC:
-
area under the curve
- BMI:
-
body mass index
- BPA:
-
bisphenol A
- BPF:
-
bisphenol F
- BPS:
-
bisphenol S
- DES:
-
diethylstilbestrol
- DMRs:
-
differentially methylated regions
- EWAS:
-
Epigenome wide association study
- GST:
-
glutathione transferase
- H3K4:
-
Histone 3 lysine 4
- HNSCC:
-
head and neck squamous cell carcinoma
- ICR:
-
imprinting control regions
- IQR:
-
inter-quartile range
- LINE:
-
long interspersed nucleotide elements
- mQTL:
-
methylation quantitative trait loci
- NTD:
-
neural tube defects
- PBMC:
-
peripheral blood mononuclear cells
- PCBs:
-
polychlorinated biphenyls
- PFASs:
-
per-and polyfluoroalkyl substances
- PM10:
-
inhalable particulate matter
- PM2.5:
-
fine particulate matter
- RRBS:
-
reduced-representation bisulfite sequencing
- SAM:
-
S-adenosyl methionine
- UV:
-
ultraviolet
References
Morgan HD, Santos F, Green K, Dean W, Reik W (2005) Epigenetic reprogramming in mammals. Hum Mol Genet 14 Spec No 1:R47–58. https://doi.org/10.1093/hmg/ddi114
Gluckman PD, Hanson MA, Cooper C, Thornburg KL (2008) Effect of in utero and early-life conditions on adult health and disease. N Engl J Med 359(1):61–73. https://doi.org/10.1056/NEJMra0708473
Li E (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 3(9):662–673. https://doi.org/10.1038/nrg887
Daly LE, Kirke PN, Molloy A, Weir DG, Scott JM (1995) Folate levels and neural tube defects. Implications for prevention JAMA 274(21):1698–1702. https://doi.org/10.1001/jama.1995.03530210052030
Dunlevy LP, Burren KA, Mills K, Chitty LS, Copp AJ, Greene ND (2006) Integrity of the methylation cycle is essential for mammalian neural tube closure. Birth Defects Res A Clin Mol Teratol 76(7):544–552. https://doi.org/10.1002/bdra.20286
Wang L, Wang F, Guan J, Le J, Wu L, Zou J, Zhao H, Pei L, Zheng X, Zhang T (2010) Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects. Am J Clin Nutr 91(5):1359–1367. https://doi.org/10.3945/ajcn.2009.28858
Yan L, Zhao L, Long Y, Zou P, Ji G, Gu A, Zhao P (2012) Association of the maternal MTHFR C677T polymorphism with susceptibility to neural tube defects in offsprings: evidence from 25 case-control studies. PLoS One 7(10):e41689. https://doi.org/10.1371/journal.pone.0041689
Waterland RA, Michels KB (2007) Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 27:363–388. https://doi.org/10.1146/annurev.nutr.27.061406.093705
Gluckman PD, Hanson MA (2004) Developmental origins of disease paradigm: a mechanistic and evolutionary perspective. Pediatr Res 56(3):311–317. https://doi.org/10.1203/01.PDR.0000135998.08025.FB
Johansson M, Relton C, Ueland PM, Vollset SE, Midttun O, Nygard O, Slimani N, Boffetta P, Jenab M, Clavel-Chapelon F, Boutron-Ruault MC, Fagherazzi G, Kaaks R, Rohrmann S, Boeing H, Weikert C, Bueno-de-Mesquita HB, Ros MM, van Gils CH, Peeters PH, Agudo A, Barricarte A, Navarro C, Rodriguez L, Sanchez MJ, Larranaga N, Khaw KT, Wareham N, Allen NE, Crowe F, Gallo V, Norat T, Krogh V, Masala G, Panico S, Sacerdote C, Tumino R, Trichopoulou A, Lagiou P, Trichopoulos D, Rasmuson T, Hallmans G, Riboli E, Vineis P, Brennan P (2010) Serum B vitamin levels and risk of lung cancer. JAMA 303(23):2377–2385. https://doi.org/10.1001/jama.2010.808
Freudenheim JL, Marshall JR, Vena JE, Laughlin R, Brasure JR, Swanson MK, Nemoto T, Graham S (1996) Premenopausal breast cancer risk and intake of vegetables, fruits, and related nutrients. J Natl Cancer Inst 88(6):340–348. https://doi.org/10.1093/jnci/88.6.340
Graham S, Hellmann R, Marshall J, Freudenheim J, Vena J, Swanson M, Zielezny M, Nemoto T, Stubbe N, Raimondo T (1991) Nutritional epidemiology of postmenopausal breast cancer in western New York. Am J Epidemiol 134(6):552–566. https://doi.org/10.1093/oxfordjournals.aje.a116129
Martinez ME, Marshall JR, Giovannucci E (2008) Diet and cancer prevention: the roles of observation and experimentation. Nat Rev Cancer 8(9):694–703. https://doi.org/10.1038/nrc2441
Kiss E, Forika G, Mohacsi R, Nemeth Z, Krenacs T, Dank M (2021) Methyl-donors can induce apoptosis and attenuate both the Akt and the Erk1/2 mediated proliferation pathways in breast and lung cancer cell lines. Int J Mol Sci 22(7). https://doi.org/10.3390/ijms22073598
Kim DH, Smith-Warner SA, Spiegelman D, Yaun SS, Colditz GA, Freudenheim JL, Giovannucci E, Goldbohm RA, Graham S, Harnack L, Jacobs EJ, Leitzmann M, Mannisto S, Miller AB, Potter JD, Rohan TE, Schatzkin A, Speizer FE, Stevens VL, Stolzenberg-Solomon R, Terry P, Toniolo P, Weijenberg MP, Willett WC, Wolk A, Zeleniuch-Jacquotte A, Hunter DJ (2010) Pooled analyses of 13 prospective cohort studies on folate intake and colon cancer. Cancer Causes Control 21(11):1919–1930. https://doi.org/10.1007/s10552-010-9620-8
Zhao Y, Guo C, Hu H, Zheng L, Ma J, Jiang L, Zhao E, Li H (2017) Folate intake, serum folate levels and esophageal cancer risk: an overall and dose-response meta-analysis. Oncotarget 8(6):10458–10469. https://doi.org/10.18632/oncotarget.14432
Qiang Y, Li Q, Xin Y, Fang X, Tian Y, Ma J, Wang J, Wang Q, Zhang R, Wang J, Wang F (2018) Intake of dietary one-carbon metabolism-related B vitamins and the risk of Esophageal cancer: a dose-response meta-analysis. Nutrients 10(7). https://doi.org/10.3390/nu10070835
de Vogel S, Wouters KA, Gottschalk RW, van Schooten FJ, de Goeij AF, de Bruine AP, Goldbohm RA, van den Brandt PA, van Engeland M, Weijenberg MP (2011) Dietary methyl donors, methyl metabolizing enzymes, and epigenetic regulators: diet-gene interactions and promoter CpG island hypermethylation in colorectal cancer. Cancer Causes Control 22(1):1–12. https://doi.org/10.1007/s10552-010-9659-6
Huang CY, Abulimiti A, Zhang X, Feng XL, Luo H, Chen YM, Fang YJ, Zhang CX (2020) Dietary B vitamin and methionine intakes and risk for colorectal cancer: a case-control study in China. Br J Nutr 123(11):1277–1289. https://doi.org/10.1017/S0007114520000501
van Driel LM, Eijkemans MJ, de Jonge R, de Vries JH, van Meurs JB, Steegers EA, Steegers-Theunissen RP (2009) Body mass index is an important determinant of methylation biomarkers in women of reproductive ages. J Nutr 139(12):2315–2321. https://doi.org/10.3945/jn.109.109710
Park HJ, Bailey LB, Shade DC, Hausman DB, Hohos NM, Meagher RB, Kauwell GP, Lewis RD, Smith AK (2017) Distinctions in gene-specific changes in DNA methylation in response to folic acid supplementation between women with normal weight and obesity. Obes Res Clin Pract 11(6):665–676. https://doi.org/10.1016/j.orcp.2017.06.004
Pauwels S, Ghosh M, Duca RC, Bekaert B, Freson K, Huybrechts I, Langie SAS, Koppen G, Devlieger R, Godderis L (2017) Maternal intake of methyl-group donors affects DNA methylation of metabolic genes in infants. Clin Epigenetics 9:16. https://doi.org/10.1186/s13148-017-0321-y
Caffrey A, Irwin RE, McNulty H, Strain JJ, Lees-Murdock DJ, McNulty BA, Ward M, Walsh CP, Pentieva K (2018) Gene-specific DNA methylation in newborns in response to folic acid supplementation during the second and third trimesters of pregnancy: epigenetic analysis from a randomized controlled trial. Am J Clin Nutr 107(4):566–575. https://doi.org/10.1093/ajcn/nqx069
Bakulski KM, Dou JF, Feinberg JI, Brieger KK, Croen LA, Hertz-Picciotto I, Newschaffer CJ, Schmidt RJ, Fallin MD (2020) Prenatal multivitamin use and MTHFR genotype are associated with Newborn cord blood DNA methylation. Int J Environ Res Public Health 17(24). https://doi.org/10.3390/ijerph17249190
Ingrosso D, Cimmino A, Perna AF, Masella L, De Santo NG, De Bonis ML, Vacca M, D’Esposito M, D’Urso M, Galletti P, Zappia V (2003) Folate treatment and unbalanced methylation and changes of allelic expression induced by hyperhomocysteinaemia in patients with uraemia. Lancet 361(9370):1693–1699. https://doi.org/10.1016/S0140-6736(03)13372-7
Pufulete M, Al-Ghnaniem R, Leather AJ, Appleby P, Gout S, Terry C, Emery PW, Sanders TA (2003) Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology 124(5):1240–1248. https://doi.org/10.1016/s0016-5085(03)00279-8
Pufulete M, Al-Ghnaniem R, Rennie JA, Appleby P, Harris N, Gout S, Emery PW, Sanders TA (2005) Influence of folate status on genomic DNA methylation in colonic mucosa of subjects without colorectal adenoma or cancer. Br J Cancer 92(5):838–842. https://doi.org/10.1038/sj.bjc.6602439
Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, Kelsey KT (2007) Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomark Prev 16(1):108–114. https://doi.org/10.1158/1055-9965.EPI-06-0636
Frederick AM, Guo C, Meyer A, Yan L, Schneider SS, Liu Z (2021) The influence of obesity on folate status, DNA methylation and cancer-related gene expression in normal breast tissues from premenopausal women. Epigenetics 16(4):458–467. https://doi.org/10.1080/15592294.2020.1805687
Song MA, Brasky TM, Marian C, Weng DY, Taslim C, Llanos AA, Dumitrescu RG, Liu Z, Mason JB, Spear SL, Kallakury BV, Freudenheim JL, Shields PG (2016) Genetic variation in one-carbon metabolism in relation to genome-wide DNA methylation in breast tissue from heathy women. Carcinogenesis 37(5):471–480. https://doi.org/10.1093/carcin/bgw030
Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS (2009) Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol 5(7):401–408. https://doi.org/10.1038/nrendo.2009.102
Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW (2010) DNA methylation as a biomarker for cardiovascular disease risk. PLoS One 5(3):e9692. https://doi.org/10.1371/journal.pone.0009692
Papait R, Condorelli G (2010) Epigenetics in heart failure. Ann N Y Acad Sci 1188:159–164. https://doi.org/10.1111/j.1749-6632.2009.05096.x
Christensen BC, Kelsey KT, Zheng S, Houseman EA, Marsit CJ, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Kushi LH, Kwan ML, Wiencke JK (2010) Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet 6(7):e1001043. https://doi.org/10.1371/journal.pgen.1001043
Sanchez H, Hossain MB, Lera L, Hirsch S, Albala C, Uauy R, Broberg K, Ronco AM (2017) High levels of circulating folate concentrations are associated with DNA methylation of tumor suppressor and repair genes p16, MLH1, and MGMT in elderly Chileans. Clin Epigenetics 9:74. https://doi.org/10.1186/s13148-017-0374-y
Yeh CC, Goyal A, Shen J, Wu HC, Strauss JA, Wang Q, Gurvich I, Safyan RA, Manji GA, Gamble MV, Siegel AB, Santella RM (2017) Global level of plasma DNA methylation is associated with overall survival in patients with hepatocellular carcinoma. Ann Surg Oncol 24(12):3788–3795. https://doi.org/10.1245/s10434-017-5913-4
Li Q, Ding L, Jing N, Liu C, Yang Z, Chen F, Hou L, Wang J (2018) Folate deficiency and aberrant DNA methylation and expression of FHIT gene were associated with cervical pathogenesis. Oncol Lett 15(2):1963–1972. https://doi.org/10.3892/ol.2017.7471
Hathcock JN (1997) Vitamins and minerals: efficacy and safety. Am J Clin Nutr 66(2):427–437. https://doi.org/10.1093/ajcn/66.2.427
Sauer J, Mason JB, Choi SW (2009) Too much folate: a risk factor for cancer and cardiovascular disease? Curr Opin Clin Nutr Metab Care 12(1):30–36. https://doi.org/10.1097/MCO.0b013e32831cec62
Tchantchou F, Graves M, Falcone D, Shea TB (2008) S-adenosylmethionine mediates glutathione efficacy by increasing glutathione S-transferase activity: implications for S-adenosyl methionine as a neuroprotective dietary supplement. J Alzheimers Dis 14(3):323–328. https://doi.org/10.3233/jad-2008-14306
Hillman RS, Steinberg SE (1982) The effects of alcohol on folate metabolism. Annu Rev Med 33:345–354. https://doi.org/10.1146/annurev.me.33.020182.002021
Dumitrescu RG, Shields PG (2005) The etiology of alcohol-induced breast cancer. Alcohol 35(3):213–225. https://doi.org/10.1016/j.alcohol.2005.04.005
Kharbanda KK (2009) Alcoholic liver disease and methionine metabolism. Semin Liver Dis 29(2):155–165. https://doi.org/10.1055/s-0029-1214371
Alcohol drinking. IARC Working Group, Lyon, 13–20 October 1987 (1988) IARC Monogr Eval Carcinog Risks Hum 44:1–378
Giovannucci E, Rimm EB, Ascherio A, Stampfer MJ, Colditz GA, Willett WC (1995) Alcohol, low-methionine – low-folate diets, and risk of colon cancer in men. J Natl Cancer Inst 87 (4):265–273. https://doi.org/10.1093/jnci/87.4.265
Schernhammer ES, Giovannucci E, Kawasaki T, Rosner B, Fuchs CS, Ogino S (2010) Dietary folate, alcohol and B vitamins in relation to LINE-1 hypomethylation in colon cancer. Gut 59(6):794–799. https://doi.org/10.1136/gut.2009.183707
Liu C, Marioni RE, Hedman AK, Pfeiffer L, Tsai PC, Reynolds LM, Just AC, Duan Q, Boer CG, Tanaka T, Elks CE, Aslibekyan S, Brody JA, Kuhnel B, Herder C, Almli LM, Zhi D, Wang Y, Huan T, Yao C, Mendelson MM, Joehanes R, Liang L, Love SA, Guan W, Shah S, McRae AF, Kretschmer A, Prokisch H, Strauch K, Peters A, Visscher PM, Wray NR, Guo X, Wiggins KL, Smith AK, Binder EB, Ressler KJ, Irvin MR, Absher DM, Hernandez D, Ferrucci L, Bandinelli S, Lohman K, Ding J, Trevisi L, Gustafsson S, Sandling JH, Stolk L, Uitterlinden AG, Yet I, Castillo-Fernandez JE, Spector TD, Schwartz JD, Vokonas P, Lind L, Li Y, Fornage M, Arnett DK, Wareham NJ, Sotoodehnia N, Ong KK, van Meurs JBJ, Conneely KN, Baccarelli AA, Deary IJ, Bell JT, North KE, Liu Y, Waldenberger M, London SJ, Ingelsson E, Levy D (2018) A DNA methylation biomarker of alcohol consumption. Mol Psychiatry 23(2):422–433. https://doi.org/10.1038/mp.2016.192
Smith IM, Mydlarz WK, Mithani SK, Califano JA (2007) DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer 121(8):1724–1728. https://doi.org/10.1002/ijc.22889
Marsit CJ, Christensen BC, Houseman EA, Karagas MR, Wrensch MR, Yeh RF, Nelson HH, Wiemels JL, Zheng S, Posner MR, McClean MD, Wiencke JK, Kelsey KT (2009) Epigenetic profiling reveals etiologically distinct patterns of DNA methylation in head and neck squamous cell carcinoma. Carcinogenesis 30(3):416–422. https://doi.org/10.1093/carcin/bgp006
Hamajima N, Hirose K, Tajima K, Rohan T, Calle EE, Heath CW, Jr., Coates RJ, Liff JM, Talamini R, Chantarakul N, Koetsawang S, Rachawat D, Morabia A, Schuman L, Stewart W, Szklo M, Bain C, Schofield F, Siskind V, Band P, Coldman AJ, Gallagher RP, Hislop TG, Yang P, Kolonel LM, Nomura AM, Hu J, Johnson KC, Mao Y, De Sanjose S, Lee N, Marchbanks P, Ory HW, Peterson HB, Wilson HG, Wingo PA, Ebeling K, Kunde D, Nishan P, Hopper JL, Colditz G, Gajalanski V, Martin N, Pardthaisong T, Silpisornkosol S, Theetranont C, Boosiri B, Chutivongse S, Jimakorn P, Virutamasen P, Wongsrichanalai C, Ewertz M, Adami HO, Bergkvist L, Magnusson C, Persson I, Chang-Claude J, Paul C, Skegg DC, Spears GF, Boyle P, Evstifeeva T, Daling JR, Hutchinson WB, Malone K, Noonan EA, Stanford JL, Thomas DB, Weiss NS, White E, Andrieu N, Bremond A, Clavel F, Gairard B, Lansac J, Piana L, Renaud R, Izquierdo A, Viladiu P, Cuevas HR, Ontiveros P, Palet A, Salazar SB, Aristizabel N, Cuadros A, Tryggvadottir L, Tulinius H, Bachelot A, Le MG, Peto J, Franceschi S, Lubin F, Modan B, Ron E, Wax Y, Friedman GD, Hiatt RA, Levi F, Bishop T, Kosmelj K, Primic-Zakelj M, Ravnihar B, Stare J, Beeson WL, Fraser G, Bullbrook RD, Cuzick J, Duffy SW, Fentiman IS, Hayward JL, Wang DY, McMichael AJ, McPherson K, Hanson RL, Leske MC, Mahoney MC, Nasca PC, Varma AO, Weinstein AL, Moller TR, Olsson H, Ranstam J, Goldbohm RA, van den Brandt PA, Apelo RA, Baens J, de la Cruz JR, Javier B, Lacaya LB, Ngelangel CA, La Vecchia C, Negri E, Marubini E, Ferraroni M, Gerber M, Richardson S, Segala C, Gatei D, Kenya P, Kungu A, Mati JG, Brinton LA, Hoover R, Schairer C, Spirtas R, Lee HP, Rookus MA, van Leeuwen FE, Schoenberg JA, McCredie M, Gammon MD, Clarke EA, Jones L, Neil A, Vessey M, Yeates D, Appleby P, Banks E, Beral V, Bull D, Crossley B, Goodill A, Green J, Hermon C, Key T, Langston N, Lewis C, Reeves G, Collins R, Doll R, Peto R, Mabuchi K, Preston D, Hannaford P, Kay C, Rosero-Bixby L, Gao YT, Jin F, Yuan JM, Wei HY, Yun T, Zhiheng C, Berry G, Cooper Booth J, Jelihovsky T, MacLennan R, Shearman R, Wang QS, Baines CJ, Miller AB, Wall C, Lund E, Stalsberg H, Shu XO, Zheng W, Katsouyanni K, Trichopoulou A, Trichopoulos D, Dabancens A, Martinez L, Molina R, Salas O, Alexander FE, Anderson K, Folsom AR, Hulka BS, Bernstein L, Enger S, Haile RW, Paganini-Hill A, Pike MC, Ross RK, Ursin G, Yu MC, Longnecker MP, Newcomb P, Bergkvist L, Kalache A, Farley TM, Holck S, Meirik O, Collaborative Group on Hormonal Factors in Breast C (2002) Alcohol, tobacco and breast cancer – collaborative reanalysis of individual data from 53 epidemiological studies, including 58,515 women with breast cancer and 95,067 women without the disease. Br J Cancer 87(11):1234–1245. https://doi.org/10.1038/sj.bjc.6600596
Key J, Hodgson S, Omar RZ, Jensen TK, Thompson SG, Boobis AR, Davies DS, Elliott P (2006) Meta-analysis of studies of alcohol and breast cancer with consideration of the methodological issues. Cancer Causes Control 17(6):759–770. https://doi.org/10.1007/s10552-006-0011-0
Llanos AA, Dumitrescu RG, Brasky TM, Liu Z, Mason JB, Marian C, Makambi KH, Spear SL, Kallakury BV, Freudenheim JL, Shields PG (2015) Relationships among folate, alcohol consumption, gene variants in one-carbon metabolism and p16INK4a methylation and expression in healthy breast tissues. Carcinogenesis 36(1):60–67. https://doi.org/10.1093/carcin/bgu219
Wilson LE, Xu Z, Harlid S, White AJ, Troester MA, Sandler DP, Taylor JA (2019) Alcohol and DNA methylation: an epigenome-wide association study in blood and Normal breast tissue. Am J Epidemiol 188(6):1055–1065. https://doi.org/10.1093/aje/kwz032
Lu M, Xueying Q, Hexiang P, Wenjing G, Hagg S, Weihua C, Chunxiao L, Canqing Y, Jun L, Zengchang P, Liming C, Hua W, Xianping W, Yunzhang W, Liming L (2021) Genome-wide associations between alcohol consumption and blood DNA methylation: evidence from twin study. Epigenomics 13(12):939–951. https://doi.org/10.2217/epi-2021-0039
Kim DS, Kim YH, Lee WK, Na YK, Hong HS (2016) Effect of alcohol consumption on peripheral blood Alu methylation in Korean men. Biomarkers 21(3):243–248. https://doi.org/10.3109/1354750X.2015.1134661
Zhang H, Herman AI, Kranzler HR, Anton RF, Zhao H, Zheng W, Gelernter J (2013) Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res 37(Suppl 1):E108–E115. https://doi.org/10.1111/j.1530-0277.2012.01928.x
Philibert RA, Penaluna B, White T, Shires S, Gunter T, Liesveld J, Erwin C, Hollenbeck N, Osborn T (2014) A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics 9(9):1212–1219. https://doi.org/10.4161/epi.32252
Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 102(30):10604–10609. https://doi.org/10.1073/pnas.0500398102
Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT (2009) Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 5(8):e1000602. https://doi.org/10.1371/journal.pgen.1000602
Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, Savage DA, Mueller-Holzner E, Marth C, Kocjan G, Gayther SA, Jones A, Beck S, Wagner W, Laird PW, Jacobs IJ, Widschwendter M (2010) Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res 20(4):440–446. https://doi.org/10.1101/gr.103606.109
Horvath S (2013) DNA methylation age of human tissues and cell types. Genome Biol 14(10):R115. https://doi.org/10.1186/gb-2013-14-10-r115
Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K (2013) Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 49(2):359–367. https://doi.org/10.1016/j.molcel.2012.10.016
Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, Whitsel EA, Wilson JG, Reiner AP, Aviv A, Lohman K, Liu Y, Ferrucci L, Horvath S (2018) An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 10(4):573–591. https://doi.org/10.18632/aging.101414
Ward-Caviness CK, Nwanaji-Enwerem JC, Wolf K, Wahl S, Colicino E, Trevisi L, Kloog I, Just AC, Vokonas P, Cyrys J, Gieger C, Schwartz J, Baccarelli AA, Schneider A, Peters A (2016) Long-term exposure to air pollution is associated with biological aging. Oncotarget 7(46):74510–74525. https://doi.org/10.18632/oncotarget.12903
Nwanaji-Enwerem JC, Colicino E, Trevisi L, Kloog I, Just AC, Shen J, Brennan K, Dereix A, Hou L, Vokonas P, Schwartz J, Baccarelli AA (2016) Long-term ambient particle exposures and blood DNA methylation age: findings from the VA normative aging study. Environ Epigenet 2(2). https://doi.org/10.1093/eep/dvw006
Nwanaji-Enwerem JC, Dai L, Colicino E, Oulhote Y, Di Q, Kloog I, Just AC, Hou L, Vokonas P, Baccarelli AA, Weisskopf MG, Schwartz JD (2017) Associations between long-term exposure to PM2.5 component species and blood DNA methylation age in the elderly: the VA normative aging study. Environ Int 102:57–65. https://doi.org/10.1016/j.envint.2016.12.024
Dhingra R, Nwanaji-Enwerem JC, Samet M, Ward-Caviness CK (2018) DNA methylation age-environmental influences, health impacts, and its role in environmental epidemiology. Curr Environ Health Rep 5(3):317–327. https://doi.org/10.1007/s40572-018-0203-2
Min JL, Hemani G, Hannon E, Dekkers KF, Castillo-Fernandez J, Luijk R, Carnero-Montoro E, Lawson DJ, Burrows K, Suderman M, Bretherick AD, Richardson TG, Klughammer J, Iotchkova V, Sharp G, Al Khleifat A, Shatunov A, Iacoangeli A, McArdle WL, Ho KM, Kumar A, Soderhall C, Soriano-Tarraga C, Giralt-Steinhauer E, Kazmi N, Mason D, McRae AF, Corcoran DL, Sugden K, Kasela S, Cardona A, Day FR, Cugliari G, Viberti C, Guarrera S, Lerro M, Gupta R, Bollepalli S, Mandaviya P, Zeng Y, Clarke TK, Walker RM, Schmoll V, Czamara D, Ruiz-Arenas C, Rezwan FI, Marioni RE, Lin T, Awaloff Y, Germain M, Aissi D, Zwamborn R, van Eijk K, Dekker A, van Dongen J, Hottenga JJ, Willemsen G, Xu CJ, Barturen G, Catala-Moll F, Kerick M, Wang C, Melton P, Elliott HR, Shin J, Bernard M, Yet I, Smart M, Gorrie-Stone T, Consortium B, Shaw C, Al Chalabi A, Ring SM, Pershagen G, Melen E, Jimenez-Conde J, Roquer J, Lawlor DA, Wright J, Martin NG, Montgomery GW, Moffitt TE, Poulton R, Esko T, Milani L, Metspalu A, Perry JRB, Ong KK, Wareham NJ, Matullo G, Sacerdote C, Panico S, Caspi A, Arseneault L, Gagnon F, Ollikainen M, Kaprio J, Felix JF, Rivadeneira F, Tiemeier H, van IMH, Uitterlinden AG, Jaddoe VWV, Haley C, McIntosh AM, Evans KL, Murray A, Raikkonen K, Lahti J, Nohr EA, Sorensen TIA, Hansen T, Morgen CS, Binder EB, Lucae S, Gonzalez JR, Bustamante M, Sunyer J, Holloway JW, Karmaus W, Zhang H, Deary IJ, Wray NR, Starr JM, Beekman M, van Heemst D, Slagboom PE, Morange PE, Tregouet DA, Veldink JH, Davies GE, de Geus EJC, Boomsma DI, Vonk JM, Brunekreef B, Koppelman GH, Alarcon-Riquelme ME, Huang RC, Pennell CE, van Meurs J, Ikram MA, Hughes AD, Tillin T, Chaturvedi N, Pausova Z, Paus T, Spector TD, Kumari M, Schalkwyk LC, Visscher PM, Davey Smith G, Bock C, Gaunt TR, Bell JT, Heijmans BT, Mill J, Relton CL (2021) Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet 53(9):1311–1321. https://doi.org/10.1038/s41588-021-00923-x
Bell CG, Lowe R, Adams PD, Baccarelli AA, Beck S, Bell JT, Christensen BC, Gladyshev VN, Heijmans BT, Horvath S, Ideker T, Issa JJ, Kelsey KT, Marioni RE, Reik W, Relton CL, Schalkwyk LC, Teschendorff AE, Wagner W, Zhang K, Rakyan VK (2019) DNA methylation aging clocks: challenges and recommendations. Genome Biol 20(1):249. https://doi.org/10.1186/s13059-019-1824-y
Divine KK, Pulling LC, Marron-Terada PG, Liechty KC, Kang T, Schwartz AG, Bocklage TJ, Coons TA, Gilliland FD, Belinsky SA (2005) Multiplicity of abnormal promoter methylation in lung adenocarcinomas from smokers and never smokers. Int J Cancer 114(3):400–405. https://doi.org/10.1002/ijc.20761
Hou M, Morishita Y, Iljima T, Inadome Y, Mase K, Dai Y, Noguchi M (1999) DNA methylation and expression of p16(INK4A) gene in pulmonary adenocarcinoma and anthracosis in background lung. Int J Cancer 84(6):609–613. https://doi.org/10.1002/(sici)1097-0215(19991222)84:6<609::aid-ijc12>3.0.co;2-q
Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, Mark EJ, Kelsey KT (2001) p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res 61(8):3419–3424
Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, Virmani AK, Zochbauer-Muller S, Tsukuda K, Sugio K, Shimizu N, Shimizu K, Lee H, Chen CY, Fong KM, Gilcrease M, Roth JA, Minna JD, Gazdar AF (2003) Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer 103(2):153–160. https://doi.org/10.1002/ijc.10787
Kim DH, Kim JS, Ji YI, Shim YM, Kim H, Han J, Park J (2003) Hypermethylation of RASSF1A promoter is associated with the age at starting smoking and a poor prognosis in primary non-small cell lung cancer. Cancer Res 63(13):3743–3746
Marsit CJ, Kim DH, Liu M, Hinds PW, Wiencke JK, Nelson HH, Kelsey KT (2005) Hypermethylation of RASSF1A and BLU tumor suppressor genes in non-small cell lung cancer: implications for tobacco smoking during adolescence. Int J Cancer 114(2):219–223. https://doi.org/10.1002/ijc.20714
Wiencke JK, Kelsey KT (2002) Teen smoking, field cancerization, and a “critical period” hypothesis for lung cancer susceptibility. Environ Health Perspect 110(6):555–558. https://doi.org/10.1289/ehp.02110555
Hasegawa M, Nelson HH, Peters E, Ringstrom E, Posner M, Kelsey KT (2002) Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene 21(27):4231–4236. https://doi.org/10.1038/sj.onc.1205528
Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG (1998) Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A 95(20):11891–11896. https://doi.org/10.1073/pnas.95.20.11891
Kersting M, Friedl C, Kraus A, Behn M, Pankow W, Schuermann M (2000) Differential frequencies of p16(INK4a) promoter hypermethylation, p53 mutation, and K-ras mutation in exfoliative material mark the development of lung cancer in symptomatic chronic smokers. J Clin Oncol 18(18):3221–3229. https://doi.org/10.1200/JCO.2000.18.18.3221
Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA (2000) Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res 60(21):5954–5958
Sozzi G, Pastorino U, Moiraghi L, Tagliabue E, Pezzella F, Ghirelli C, Tornielli S, Sard L, Huebner K, Pierotti MA, Croce CM, Pilotti S (1998) Loss of FHIT function in lung cancer and preinvasive bronchial lesions. Cancer Res 58(22):5032–5037
von Zeidler SV, Miracca EC, Nagai MA, Birman EG (2004) Hypermethylation of the p16 gene in normal oral mucosa of smokers. Int J Mol Med 14(5):807–811. https://doi.org/10.3892/ijmm.14.5.807
Zochbauer-Muller S, Fong KM, Maitra A, Lam S, Geradts J, Ashfaq R, Virmani AK, Milchgrub S, Gazdar AF, Minna JD (2001) 5’ CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res 61(9):3581–3585
Zochbauer-Muller S, Lam S, Toyooka S, Virmani AK, Toyooka KO, Seidl S, Minna JD, Gazdar AF (2003) Aberrant methylation of multiple genes in the upper aerodigestive tract epithelium of heavy smokers. Int J Cancer 107(4):612–616. https://doi.org/10.1002/ijc.11458
Kulkarni V, Saranath D (2004) Concurrent hypermethylation of multiple regulatory genes in chewing tobacco associated oral squamous cell carcinomas and adjacent normal tissues. Oral Oncol 40(2):145–153. https://doi.org/10.1016/s1368-8375(03)00143-x
Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H (2011) Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet 88(4):450–457. https://doi.org/10.1016/j.ajhg.2011.03.003
Breitling LP, Salzmann K, Rothenbacher D, Burwinkel B, Brenner H (2012) Smoking, F2RL3 methylation, and prognosis in stable coronary heart disease. Eur Heart J 33(22):2841–2848. https://doi.org/10.1093/eurheartj/ehs091
Monick MM, Beach SR, Plume J, Sears R, Gerrard M, Brody GH, Philibert RA (2012) Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet B Neuropsychiatr Genet 159B(2):141–151. https://doi.org/10.1002/ajmg.b.32021
Fasanelli F, Baglietto L, Ponzi E, Guida F, Campanella G, Johansson M, Grankvist K, Johansson M, Assumma MB, Naccarati A, Chadeau-Hyam M, Ala U, Faltus C, Kaaks R, Risch A, De Stavola B, Hodge A, Giles GG, Southey MC, Relton CL, Haycock PC, Lund E, Polidoro S, Sandanger TM, Severi G, Vineis P (2015) Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat Commun 6:10192. https://doi.org/10.1038/ncomms10192
Reynolds LM, Wan M, Ding J, Taylor JR, Lohman K, Su D, Bennett BD, Porter DK, Gimple R, Pittman GS, Wang X, Howard TD, Siscovick D, Psaty BM, Shea S, Burke GL, Jacobs DR Jr, Rich SS, Hixson JE, Stein JH, Stunnenberg H, Barr RG, Kaufman JD, Post WS, Hoeschele I, Herrington DM, Bell DA, Liu Y (2015) DNA methylation of the aryl hydrocarbon receptor repressor associations with cigarette smoking and subclinical atherosclerosis. Circ Cardiovasc Genet 8(5):707–716. https://doi.org/10.1161/CIRCGENETICS.115.001097
Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P, Flanagan JM (2013) Epigenome-wide association study in the European prospective investigation into cancer and nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet 22(5):843–851. https://doi.org/10.1093/hmg/dds488
Zeilinger S, Kuhnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, Strauch K, Waldenberger M, Illig T (2013) Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One 8(5):e63812. https://doi.org/10.1371/journal.pone.0063812
Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86. https://doi.org/10.1186/1471-2105-13-86
Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, Soderhall C, Scheynius A, Kere J (2012) Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 7(7):e41361. https://doi.org/10.1371/journal.pone.0041361
Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, Guan W, Xu T, Elks CE, Aslibekyan S, Moreno-Macias H, Smith JA, Brody JA, Dhingra R, Yousefi P, Pankow JS, Kunze S, Shah SH, McRae AF, Lohman K, Sha J, Absher DM, Ferrucci L, Zhao W, Demerath EW, Bressler J, Grove ML, Huan T, Liu C, Mendelson MM, Yao C, Kiel DP, Peters A, Wang-Sattler R, Visscher PM, Wray NR, Starr JM, Ding J, Rodriguez CJ, Wareham NJ, Irvin MR, Zhi D, Barrdahl M, Vineis P, Ambatipudi S, Uitterlinden AG, Hofman A, Schwartz J, Colicino E, Hou L, Vokonas PS, Hernandez DG, Singleton AB, Bandinelli S, Turner ST, Ware EB, Smith AK, Klengel T, Binder EB, Psaty BM, Taylor KD, Gharib SA, Swenson BR, Liang L, DeMeo DL, O’Connor GT, Herceg Z, Ressler KJ, Conneely KN, Sotoodehnia N, Kardia SL, Melzer D, Baccarelli AA, van Meurs JB, Romieu I, Arnett DK, Ong KK, Liu Y, Waldenberger M, Deary IJ, Fornage M, Levy D, London SJ (2016) Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet 9(5):436–447. https://doi.org/10.1161/CIRCGENETICS.116.001506
Bauer M, Fink B, Thurmann L, Eszlinger M, Herberth G, Lehmann I (2015) Tobacco smoking differently influences cell types of the innate and adaptive immune system-indications from CpG site methylation. Clin Epigenetics 7:83. https://doi.org/10.1186/s13148-016-0249-7
Su D, Wang X, Campbell MR, Porter DK, Pittman GS, Bennett BD, Wan M, Englert NA, Crowl CL, Gimple RN, Adamski KN, Huang Z, Murphy SK, Bell DA (2016) Distinct epigenetic effects of tobacco smoking in whole blood and among leukocyte subtypes. PLoS One 11(12):e0166486. https://doi.org/10.1371/journal.pone.0166486
Flom JD, Ferris JS, Liao Y, Tehranifar P, Richards CB, Cho YH, Gonzalez K, Santella RM, Terry MB (2011) Prenatal smoke exposure and genomic DNA methylation in a multiethnic birth cohort. Cancer Epidemiol Biomark Prev 20(12):2518–2523. https://doi.org/10.1158/1055-9965.EPI-11-0553
Breton CV, Siegmund KD, Joubert BR, Wang X, Qui W, Carey V, Nystad W, Haberg SE, Ober C, Nicolae D, Barnes KC, Martinez F, Liu A, Lemanske R, Strunk R, Weiss S, London S, Gilliland F, Raby B, Bc A (2014) Prenatal tobacco smoke exposure is associated with childhood DNA CpG methylation. PLoS One 9(6):e99716. https://doi.org/10.1371/journal.pone.0099716
Lee KW, Richmond R, Hu P, French L, Shin J, Bourdon C, Reischl E, Waldenberger M, Zeilinger S, Gaunt T, McArdle W, Ring S, Woodward G, Bouchard L, Gaudet D, Smith GD, Relton C, Paus T, Pausova Z (2015) Prenatal exposure to maternal cigarette smoking and DNA methylation: epigenome-wide association in a discovery sample of adolescents and replication in an independent cohort at birth through 17 years of age. Environ Health Perspect 123(2):193–199. https://doi.org/10.1289/ehp.1408614
Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, Ring SM, Smith AD, Timpson NJ, Tilling K, Davey Smith G, Relton CL (2015) Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon longitudinal study of parents and children (ALSPAC). Hum Mol Genet 24(8):2201–2217. https://doi.org/10.1093/hmg/ddu739
Sikdar S, Joehanes R, Joubert BR, Xu CJ, Vives-Usano M, Rezwan FI, Felix JF, Ward JM, Guan W, Richmond RC, Brody JA, Kupers LK, Baiz N, Haberg SE, Smith JA, Reese SE, Aslibekyan S, Hoyo C, Dhingra R, Markunas CA, Xu T, Reynolds LM, Just AC, Mandaviya PR, Ghantous A, Bennett BD, Wang T, Consortium TB, Bakulski KM, Melen E, Zhao S, Jin J, Herceg Z, Meurs JV, Taylor JA, Baccarelli AA, Murphy SK, Liu Y, Munthe-Kaas MC, Deary IJ, Nystad W, Waldenberger M, Annesi-Maesano I, Conneely K, Jaddoe VW, Arnett D, Snieder H, Kardia SL, Relton CL, Ong KK, Ewart S, Moreno-Macias H, Romieu I, Sotoodehnia N, Fornage M, Motsinger-Reif A, Koppelman GH, Bustamante M, Levy D, London SJ (2019) Comparison of smoking-related DNA methylation between newborns from prenatal exposure and adults from personal smoking. Epigenomics 11(13):1487–1500. https://doi.org/10.2217/epi-2019-0066
Rauschert S, Melton PE, Burdge G, Craig JM, Godfrey KM, Holbrook JD, Lillycrop K, Mori TA, Beilin LJ, Oddy WH, Pennell C, Huang RC (2019) Maternal smoking during pregnancy induces persistent epigenetic changes into adolescence, independent of postnatal smoke exposure and is associated with Cardiometabolic risk. Front Genet 10:770. https://doi.org/10.3389/fgene.2019.00770
Rauschert S, Melton PE, Heiskala A, Karhunen V, Burdge G, Craig JM, Godfrey KM, Lillycrop K, Mori TA, Beilin LJ, Oddy WH, Pennell C, Jarvelin MR, Sebert S, Huang RC (2020) Machine learning-based DNA methylation score for Fetal exposure to maternal smoking: development and validation in samples collected from adolescents and adults. Environ Health Perspect 128(9):97003. https://doi.org/10.1289/EHP6076
Herbst AL, Ulfelder H, Poskanzer DC (1971) Adenocarcinoma of the vagina. Association of maternal stilbestrol therapy with tumor appearance in young women. N Engl J Med:284 (15):878-881. https://doi.org/10.1056/NEJM197104222841604
Bromer JG, Wu J, Zhou Y, Taylor HS (2009) Hypermethylation of homeobox A10 by in utero diethylstilbestrol exposure: an epigenetic mechanism for altered developmental programming. Endocrinology 150(7):3376–3382. https://doi.org/10.1210/en.2009-0071
Newbold RR (2004) Lessons learned from perinatal exposure to diethylstilbestrol. Toxicol Appl Pharmacol 199(2):142–150. https://doi.org/10.1016/j.taap.2003.11.033
Ruden DM, Xiao L, Garfinkel MD, Lu X (2005) Hsp90 and environmental impacts on epigenetic states: a model for the trans-generational effects of diethylstilbestrol on uterine development and cancer. Hum Mol Genet 14 Spec No 1:R149–155. https://doi.org/10.1093/hmg/ddi103
Titus L, Hatch EE, Drake KM, Parker SE, Hyer M, Palmer JR, Strohsnitter WC, Adam E, Herbst AL, Huo D, Hoover RN, Troisi R (2019) Reproductive and hormone-related outcomes in women whose mothers were exposed in utero to diethylstilbestrol (DES): a report from the US National Cancer Institute DES third generation study. Reprod Toxicol 84:32–38. https://doi.org/10.1016/j.reprotox.2018.12.008
Kioumourtzoglou MA, Coull BA, O’Reilly EJ, Ascherio A, Weisskopf MG (2018) Association of Exposure to Diethylstilbestrol during pregnancy with multigenerational neurodevelopmental deficits. JAMA Pediatr 172(7):670–677. https://doi.org/10.1001/jamapediatrics.2018.0727
vom Saal FS, Hughes C (2005) An extensive new literature concerning low-dose effects of bisphenol a shows the need for a new risk assessment. Environ Health Perspect 113(8):926–933. https://doi.org/10.1289/ehp.7713
Schonfelder G, Wittfoht W, Hopp H, Talsness CE, Paul M, Chahoud I (2002) Parent bisphenol a accumulation in the human maternal-fetal-placental unit. Environ Health Perspect 110(11):A703–A707. https://doi.org/10.1289/ehp.110-1241091
Dolinoy DC, Huang D, Jirtle RL (2007) Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A 104(32):13056–13061. https://doi.org/10.1073/pnas.0703739104
Avissar-Whiting M, Veiga KR, Uhl KM, Maccani MA, Gagne LA, Moen EL, Marsit CJ (2010) Bisphenol a exposure leads to specific microRNA alterations in placental cells. Reprod Toxicol 29(4):401–406. https://doi.org/10.1016/j.reprotox.2010.04.004
Lehmler HJ, Liu B, Gadogbe M, Bao W (2018) Exposure to bisphenol a, bisphenol F, and bisphenol S in U.S. adults and children: the National Health and nutrition examination survey 2013–2014. ACS Omega 3(6):6523–6532. https://doi.org/10.1021/acsomega.8b00824
Nair VA, Valo S, Peltomaki P, Bajbouj K, Abdel-Rahman WM (2020) Oncogenic potential of bisphenol a and common environmental contaminants in human mammary epithelial cells. Int J Mol Sci 21(10). https://doi.org/10.3390/ijms21103735
Fatma Karaman E, Caglayan M, Sancar-Bas S, Ozal-Coskun C, Arda-Pirincci P, Ozden S (2019) Global and region-specific post-transcriptional and post-translational modifications of bisphenol a in human prostate cancer cells. Environ Pollut 255(Pt 2):113318. https://doi.org/10.1016/j.envpol.2019.113318
McCabe CF, Padmanabhan V, Dolinoy DC, Domino SE, Jones TR, Bakulski KM, Goodrich JM (2020) Maternal environmental exposure to bisphenols and epigenome-wide DNA methylation in infant cord blood. Environ Epigenet 6(1):dvaa021. https://doi.org/10.1093/eep/dvaa021
Engdahl E, Svensson K, Lin PD, Alavian-Ghavanini A, Lindh C, Ruegg J, Bornehag CG (2021) DNA methylation at GRIN2B partially mediates the association between prenatal bisphenol F exposure and cognitive functions in 7-year-old children in the SELMA study. Environ Int 156:106617. https://doi.org/10.1016/j.envint.2021.106617
Alavian-Ghavanini A, Lin PI, Lind PM, Risen Rimfors S, Halin Lejonklou M, Dunder L, Tang M, Lindh C, Bornehag CG, Ruegg J (2018) Prenatal bisphenol a exposure is linked to epigenetic changes in glutamate receptor subunit gene Grin2b in female rats and humans. Sci Rep 8(1):11315. https://doi.org/10.1038/s41598-018-29732-9
Wang Z, DeWitt JC, Higgins CP, Cousins IT (2017) A never-ending story of per- and Polyfluoroalkyl substances (PFASs)? Environ Sci Technol 51(5):2508–2518. https://doi.org/10.1021/acs.est.6b04806
Blum A, Balan SA, Scheringer M, Trier X, Goldenman G, Cousins IT, Diamond M, Fletcher T, Higgins C, Lindeman AE, Peaslee G, de Voogt P, Wang Z, Weber R (2015) The Madrid statement on poly- and Perfluoroalkyl substances (PFASs). Environ Health Perspect 123(5):A107–A111. https://doi.org/10.1289/ehp.1509934
Fenton SE, Ducatman A, Boobis A, DeWitt JC, Lau C, Ng C, Smith JS, Roberts SM (2021) Per- and Polyfluoroalkyl substance toxicity and human health review: current state of knowledge and strategies for informing future research. Environ Toxicol Chem 40(3):606–630. https://doi.org/10.1002/etc.4890
Program NT (2016) NTP monograph on immunotoxicity associated with exposure to perfluorooctanoic acid (PFOA) or perfluorooctane sulfonate (PFOS)
Liew Z, Goudarzi H, Oulhote Y (2018) Developmental exposures to Perfluoroalkyl substances (PFASs): an update of associated health outcomes. Curr Environ Health Rep 5(1):1–19. https://doi.org/10.1007/s40572-018-0173-4
Watkins DJ, Wellenius GA, Butler RA, Bartell SM, Fletcher T, Kelsey KT (2014) Associations between serum perfluoroalkyl acids and LINE-1 DNA methylation. Environ Int 63:71–76. https://doi.org/10.1016/j.envint.2013.10.018
Leter G, Consales C, Eleuteri P, Uccelli R, Specht IO, Toft G, Moccia T, Budillon A, Jonsson BA, Lindh CH, Giwercman A, Pedersen HS, Ludwicki JK, Zviezdai V, Heederik D, Bonde JP, Spano M (2014) Exposure to perfluoroalkyl substances and sperm DNA global methylation in Arctic and European populations. Environ Mol Mutagen 55(7):591–600. https://doi.org/10.1002/em.21874
van den Dungen MW, Murk AJ, Kampman E, Steegenga WT, Kok DE (2017) Association between DNA methylation profiles in leukocytes and serum levels of persistent organic pollutants in Dutch men. Environ Epigenet 3(1):dvx001. https://doi.org/10.1093/eep/dvx001
Guerrero-Preston R, Goldman LR, Brebi-Mieville P, Ili-Gangas C, Lebron C, Witter FR, Apelberg BJ, Hernandez-Roystacher M, Jaffe A, Halden RU, Sidransky D (2010) Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics 5(6):539–546. https://doi.org/10.4161/epi.5.6.12378
Liu CY, Chen PC, Lien PC, Liao YP (2018) Prenatal Perfluorooctyl sulfonate exposure and Alu DNA Hypomethylation in cord blood. Int J Environ Res Public Health 15(6). https://doi.org/10.3390/ijerph15061066
Kobayashi S, Azumi K, Goudarzi H, Araki A, Miyashita C, Kobayashi S, Itoh S, Sasaki S, Ishizuka M, Nakazawa H, Ikeno T, Kishi R (2017) Effects of prenatal perfluoroalkyl acid exposure on cord blood IGF2/H19 methylation and ponderal index: the Hokkaido study. J Expo Sci Environ Epidemiol 27(3):251–259. https://doi.org/10.1038/jes.2016.50
Kingsley SL, Kelsey KT, Butler R, Chen A, Eliot MN, Romano ME, Houseman A, Koestler DC, Lanphear BP, Yolton K, Braun JM (2017) Maternal serum PFOA concentration and DNA methylation in cord blood: a pilot study. Environ Res 158:174–178. https://doi.org/10.1016/j.envres.2017.06.013
Leung YK, Ouyang B, Niu L, Xie C, Ying J, Medvedovic M, Chen A, Weihe P, Valvi D, Grandjean P, Ho SM (2018) Identification of sex-specific DNA methylation changes driven by specific chemicals in cord blood in a Faroese birth cohort. Epigenetics 13(3):290–300. https://doi.org/10.1080/15592294.2018.1445901
Miura R, Araki A, Miyashita C, Kobayashi S, Kobayashi S, Wang SL, Chen CH, Miyake K, Ishizuka M, Iwasaki Y, Ito YM, Kubota T, Kishi R (2018) An epigenome-wide study of cord blood DNA methylations in relation to prenatal perfluoroalkyl substance exposure: the Hokkaido study. Environ Int 115:21–28. https://doi.org/10.1016/j.envint.2018.03.004
Ouidir M, Mendola P, Buck Louis GM, Kannan K, Zhang C, Tekola-Ayele F (2020) Concentrations of persistent organic pollutants in maternal plasma and epigenome-wide placental DNA methylation. Clin Epigenetics 12(1):103. https://doi.org/10.1186/s13148-020-00894-6
Sonkar R, Kay MK, Choudhury M (2019) PFOS modulates interactive epigenetic regulation in first-trimester human trophoblast cell line HTR-8/SVneo. Chem Res Toxicol 32(10):2016–2027. https://doi.org/10.1021/acs.chemrestox.9b00198
Szilagyi JT, Freedman AN, Kepper SL, Keshava AM, Bangma JT, Fry RC (2020) Per- and Polyfluoroalkyl substances differentially inhibit placental trophoblast migration and invasion in vitro. Toxicol Sci 175(2):210–219. https://doi.org/10.1093/toxsci/kfaa043
van den Dungen MW, Murk AJ, Kok DE, Steegenga WT (2017) Persistent organic pollutants alter DNA methylation during human adipocyte differentiation. Toxicol In Vitro 40:79–87. https://doi.org/10.1016/j.tiv.2016.12.011
Little JB (2000) Radiation carcinogenesis. Carcinogenesis 21(3):397–404. https://doi.org/10.1093/carcin/21.3.397
McDonald JW, Taylor JA, Watson MA, Saccomanno G, Devereux TR (1995) p53 and K-ras in radon-associated lung adenocarcinoma. Cancer Epidemiol Biomark Prev 4(7):791–793
Prise KM, Pinto M, Newman HC, Michael BD (2001) A review of studies of ionizing radiation-induced double-strand break clustering. Radiat Res 156(5 Pt 2):572–576. https://doi.org/10.1667/0033-7587(2001)156[0572:arosoi]2.0.co;2
Swafford DS, Middleton SK, Palmisano WA, Nikula KJ, Tesfaigzi J, Baylin SB, Herman JG, Belinsky SA (1997) Frequent aberrant methylation of p16INK4a in primary rat lung tumors. Mol Cell Biol 17(3):1366–1374. https://doi.org/10.1128/MCB.17.3.1366
Belinsky SA, Klinge DM, Liechty KC, March TH, Kang T, Gilliland FD, Sotnic N, Adamova G, Rusinova G, Telnov V (2004) Plutonium targets the p16 gene for inactivation by promoter hypermethylation in human lung adenocarcinoma. Carcinogenesis 25(6):1063–1067. https://doi.org/10.1093/carcin/bgh096
Romanenko A, Morell-Quadreny L, Lopez-Guerrero JA, Pellin A, Nepomnyaschy V, Vozianov A, Llombart-Bosch A (2002) P16INK4A and p15INK4B gene alteration associated with oxidative stress in renal cell carcinomas after the Chernobyl accident (pilot study). Diagn Mol Pathol 11(3):163–169. https://doi.org/10.1097/00019606-200209000-00007
Cleary HJ, Boulton E, Plumb M (1999) Allelic loss and promoter hypermethylation of the p15INK4b gene features in mouse radiation-induced lymphoid – but not myeloid – leukaemias. Leukemia 13(12):2049–2052. https://doi.org/10.1038/sj.leu.2401616
Malumbres M, Perez de Castro I, Santos J, Fernandez Piqueras J, Pellicer A (1999) Hypermethylation of the cell cycle inhibitor p15INK4b 3′-untranslated region interferes with its transcriptional regulation in primary lymphomas. Oncogene 18(2):385–396. https://doi.org/10.1038/sj.onc.1202299
Malumbres M, Perez de Castro I, Santos J, Melendez B, Mangues R, Serrano M, Pellicer A, Fernandez-Piqueras J (1997) Inactivation of the cyclin-dependent kinase inhibitor p15INK4b by deletion and de novo methylation with independence of p16INK4a alterations in murine primary T-cell lymphomas. Oncogene 14(11):1361–1370. https://doi.org/10.1038/sj.onc.1200969
Song N, Hsu CW, Pan H, Zheng Y, Hou L, Sim JA, Li Z, Mulder H, Easton J, Walker E, Neale G, Wilson CL, Ness KK, Krull KR, Srivastava DK, Yasui Y, Zhang J, Hudson MM, Robison LL, Huang IC, Wang Z (2021) Persistent variations of blood DNA methylation associated with treatment exposures and risk for cardiometabolic outcomes in long-term survivors of childhood cancer in the St. Jude Lifetime Cohort Genome Med 13(1):53. https://doi.org/10.1186/s13073-021-00875-1
Nair-Shalliker V, Dhillon V, Clements M, Armstrong BK, Fenech M (2014) The association between personal sun exposure, serum vitamin D and global methylation in human lymphocytes in a population of healthy adults in South Australia. Mutat Res 765:6–10. https://doi.org/10.1016/j.mrfmmm.2014.04.001
Page CM, Djordjilovic V, Nost TH, Ghiasvand R, Sandanger TM, Frigessi A, Thoresen M, Veierod MB (2020) Lifetime ultraviolet radiation exposure and DNA methylation in blood leukocytes: the Norwegian women and cancer study. Sci Rep 10(1):4521. https://doi.org/10.1038/s41598-020-61430-3
Rossman TG (2003) Mechanism of arsenic carcinogenesis: an integrated approach. Mutat Res 533(1–2):37–65. https://doi.org/10.1016/j.mrfmmm.2003.07.009
Mass MJ, Wang L (1997) Arsenic alters cytosine methylation patterns of the promoter of the tumor suppressor gene p53 in human lung cells: a model for a mechanism of carcinogenesis. Mutat Res 386(3):263–277. https://doi.org/10.1016/s1383-5742(97)00008-2
Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP (1997) Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci U S A 94(20):10907–10912. https://doi.org/10.1073/pnas.94.20.10907
Zhong CX, Mass MJ (2001) Both hypomethylation and hypermethylation of DNA associated with arsenite exposure in cultures of human cells identified by methylation-sensitive arbitrarily-primed PCR. Toxicol Lett 122(3):223–234. https://doi.org/10.1016/s0378-4274(01)00365-4
Marsit CJ, Karagas MR, Danaee H, Liu M, Andrew A, Schned A, Nelson HH, Kelsey KT (2006) Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis 27(1):112–116. https://doi.org/10.1093/carcin/bgi172
Okoji RS, Yu RC, Maronpot RR, Froines JR (2002) Sodium arsenite administration via drinking water increases genome-wide and ha-ras DNA hypomethylation in methyl-deficient C57BL/6J mice. Carcinogenesis 23(5):777–785. https://doi.org/10.1093/carcin/23.5.777
McDorman EW, Collins BW, Allen JW (2002) Dietary folate deficiency enhances induction of micronuclei by arsenic in mice. Environ Mol Mutagen 40(1):71–77. https://doi.org/10.1002/em.10085
Chanda S, Dasgupta UB, Guhamazumder D, Gupta M, Chaudhuri U, Lahiri S, Das S, Ghosh N, Chatterjee D (2006) DNA hypermethylation of promoter of gene p53 and p16 in arsenic-exposed people with and without malignancy. Toxicol Sci 89(2):431–437. https://doi.org/10.1093/toxsci/kfj030
Majumdar S, Chanda S, Ganguli B, Mazumder DN, Lahiri S, Dasgupta UB (2010) Arsenic exposure induces genomic hypermethylation. Environ Toxicol 25(3):315–318. https://doi.org/10.1002/tox.20497
Pilsner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Graziano JH, Gamble MV (2007) Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr 86(4):1179–1186. https://doi.org/10.1093/ajcn/86.4.1179
Gamble MV, Liu X, Slavkovich V, Pilsner JR, Ilievski V, Factor-Litvak P, Levy D, Alam S, Islam M, Parvez F, Ahsan H, Graziano JH (2007) Folic acid supplementation lowers blood arsenic. Am J Clin Nutr 86(4):1202–1209. https://doi.org/10.1093/ajcn/86.4.1202
Pilsner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Graziano JH, Gamble MV (2009) Folate deficiency, hyperhomocysteinemia, low urinary creatinine, and hypomethylation of leukocyte DNA are risk factors for arsenic-induced skin lesions. Environ Health Perspect 117(2):254–260. https://doi.org/10.1289/ehp.11872
Kile ML, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, Cardenas A, Wright RO, Christiani DC (2014) Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics 9(5):774–782. https://doi.org/10.4161/epi.28153
Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ (2013) Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ Health Perspect 121(8):971–977. https://doi.org/10.1289/ehp.1205925
Bozack AK, Cardenas A, Quamruzzaman Q, Rahman M, Mostofa G, Christiani DC, Kile ML (2018) DNA methylation in cord blood as mediator of the association between prenatal arsenic exposure and gestational age. Epigenetics 13(9):923–940. https://doi.org/10.1080/15592294.2018.1516453
DiGiovanni A, Demanelis K, Tong L, Argos M, Shinkle J, Jasmine F, Sabarinathan M, Rakibuz-Zaman M, Sarwar G, Islam MT, Shahriar H, Islam T, Rahman M, Yunus M, Graziano J, Gamble MV, Ahsan H, Pierce BL (2020) Assessing the impact of arsenic metabolism efficiency on DNA methylation using Mendelian randomization. Environ Epidemiol 4(2):e083. https://doi.org/10.1097/EE9.0000000000000083
Marsit CJ, Eddy K, Kelsey KT (2006) MicroRNA responses to cellular stress. Cancer Res 66(22):10843–10848. https://doi.org/10.1158/0008-5472.CAN-06-1894
Tumolo MR, Panico A, De Donno A, Mincarone P, Leo CG, Guarino R, Bagordo F, Serio F, Idolo A, Grassi T, Sabina S (2020) The expression of microRNAs and exposure to environmental contaminants related to human health: a review. Int J Environ Health Res:1–23. https://doi.org/10.1080/09603123.2020.1757043
Ramirez T, Brocher J, Stopper H, Hock R (2008) Sodium arsenite modulates histone acetylation, histone deacetylase activity and HMGN protein dynamics in human cells. Chromosoma 117(2):147–157. https://doi.org/10.1007/s00412-007-0133-5
Howe CG, Gamble MV (2016) Influence of arsenic on global levels of histone posttranslational modifications: a review of the literature and challenges in the field. Curr Environ Health Rep 3(3):225–237. https://doi.org/10.1007/s40572-016-0104-1
(IARC) IAfRoC (1993) Summaries & Evaluations: cadmium and cadmium compounds. Summaries & Evaluations 58
Registry AfTSaD (2019) ATSDR’s Substance Priority List. https://www.atsdr.cdc.gov/spl/index.html
Benbrahim-Tallaa L, Waterland RA, Dill AL, Webber MM, Waalkes MP (2007) Tumor suppressor gene inactivation during cadmium-induced malignant transformation of human prostate cells correlates with overexpression of de novo DNA methyltransferase. Environ Health Perspect 115(10):1454–1459. https://doi.org/10.1289/ehp.10207
Zhou ZH, Lei YX, Wang CX (2012) Analysis of aberrant methylation in DNA repair genes during malignant transformation of human bronchial epithelial cells induced by cadmium. Toxicol Sci 125(2):412–417. https://doi.org/10.1093/toxsci/kfr320
Yuan D, Ye S, Pan Y, Bao Y, Chen H, Shao C (2013) Long-term cadmium exposure leads to the enhancement of lymphocyte proliferation via down-regulating p16 by DNA hypermethylation. Mutat Res 757(2):125–131. https://doi.org/10.1016/j.mrgentox.2013.07.007
Xiao C, Liu Y, Xie C, Tu W, Xia Y, Costa M, Zhou X (2015) Cadmium induces histone H3 lysine methylation by inhibiting histone demethylase activity. Toxicol Sci 145(1):80–89. https://doi.org/10.1093/toxsci/kfv019
Liang ZZ, Zhu RM, Li YL, Jiang HM, Li RB, Tang LY, Wang Q, Ren ZF (2020) Differential epigenetic and transcriptional profile in MCF-7 breast cancer cells exposed to cadmium. Chemosphere 261:128148. https://doi.org/10.1016/j.chemosphere.2020.128148
Chandravanshi L, Shiv K, Kumar S (2021) Developmental toxicity of cadmium in infants and children: a review. Environ Anal Health Toxicol 36(1):e2021003–e2021000. https://doi.org/10.5620/eaht.2021003
Vilahur N, Vahter M, Broberg K (2015) The epigenetic effects of prenatal cadmium exposure. Curr Environ Health Rep 2(2):195–203. https://doi.org/10.1007/s40572-015-0049-9
Kippler M, Engstrom K, Mlakar SJ, Bottai M, Ahmed S, Hossain MB, Raqib R, Vahter M, Broberg K (2013) Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics 8(5):494–503. https://doi.org/10.4161/epi.24401
Everson TM, Punshon T, Jackson BP, Hao K, Lambertini L, Chen J, Karagas MR, Marsit CJ (2018) Cadmium-associated differential methylation throughout the placental genome: epigenome-wide association study of two U.S. birth cohorts. Environ Health Perspect 126(1):017010. https://doi.org/10.1289/EHP2192
Cowley M, Skaar DA, Jima DD, Maguire RL, Hudson KM, Park SS, Sorrow P, Hoyo C (2018) Effects of cadmium exposure on DNA methylation at imprinting control regions and genome-wide in mothers and Newborn children. Environ Health Perspect 126(3):037003. https://doi.org/10.1289/EHP2085
Millership SJ, Van de Pette M, Withers DJ (2019) Genomic imprinting and its effects on postnatal growth and adult metabolism. Cell Mol Life Sci 76(20):4009–4021. https://doi.org/10.1007/s00018-019-03197-z
Wilkinson LS, Davies W, Isles AR (2007) Genomic imprinting effects on brain development and function. Nat Rev Neurosci 8(11):832–843. https://doi.org/10.1038/nrn2235
Everson TM, Marable C, Deyssenroth MA, Punshon T, Jackson BP, Lambertini L, Karagas MR, Chen J, Marsit CJ (2019) Placental expression of imprinted genes, overall and in sex-specific patterns, associated with placental cadmium concentrations and birth size. Environ Health Perspect 127(5):57005. https://doi.org/10.1289/EHP4264
Vidal AC, Semenova V, Darrah T, Vengosh A, Huang Z, King K, Nye MD, Fry R, Skaar D, Maguire R, Murtha A, Schildkraut J, Murphy S, Hoyo C (2015) Maternal cadmium, iron and zinc levels, DNA methylation and birth weight. BMC Pharmacol Toxicol 16:20. https://doi.org/10.1186/s40360-015-0020-2
Xu P, Wu Z, Yang W, Wang L (2017) Dysregulation of DNA methylation and expression of imprinted genes in mouse placentas of fetal growth restriction induced by maternal cadmium exposure. Toxicology 390:109–116. https://doi.org/10.1016/j.tox.2017.08.003
Zhu J, Huang Z, Yang F, Zhu M, Cao J, Chen J, Lin Y, Guo S, Li J, Liu Z (2021) Cadmium disturbs epigenetic modification and induces DNA damage in mouse preimplantation embryos. Ecotoxicol Environ Saf 219:112306. https://doi.org/10.1016/j.ecoenv.2021.112306
Brooks SA, Fry RC (2017) Cadmium inhibits placental trophoblast cell migration via miRNA regulation of the transforming growth factor beta (TGF-beta) pathway. Food Chem Toxicol 109(Pt 1):721–726. https://doi.org/10.1016/j.fct.2017.07.059
Brooks SA, Martin E, Smeester L, Grace MR, Boggess K, Fry RC (2016) miRNAs as common regulators of the transforming growth factor (TGF)-beta pathway in the preeclamptic placenta and cadmium-treated trophoblasts: links between the environment, the epigenome and preeclampsia. Food Chem Toxicol 98(Pt A):50–57. https://doi.org/10.1016/j.fct.2016.06.023
Yang K, Julan L, Rubio F, Sharma A, Guan H (2006) Cadmium reduces 11 beta-hydroxysteroid dehydrogenase type 2 activity and expression in human placental trophoblast cells. Am J Physiol Endocrinol Metab 290(1):E135–E142. https://doi.org/10.1152/ajpendo.00356.2005
Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, Pegoraro V, Motta V, Tarantini L, Cantone L, Schwartz J, Bertazzi PA, Baccarelli A (2010) Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect 118(6):763–768. https://doi.org/10.1289/ehp.0901300
Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, Hu H, Sparrow D, Vokonas P, Baccarelli A (2010) Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect 118(6):790–795. https://doi.org/10.1289/ehp.0901429
Pilsner JR, Hu H, Ettinger A, Sanchez BN, Wright RO, Cantonwine D, Lazarus A, Lamadrid-Figueroa H, Mercado-Garcia A, Tellez-Rojo MM, Hernandez-Avila M (2009) Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect 117(9):1466–1471. https://doi.org/10.1289/ehp.0800497
Wu S, Hivert MF, Cardenas A, Zhong J, Rifas-Shiman SL, Agha G, Colicino E, Just AC, Amarasiriwardena C, Lin X, Litonjua AA, DeMeo DL, Gillman MW, Wright RO, Oken E, Baccarelli AA (2017) Exposure to Low levels of Lead in utero and umbilical cord blood DNA methylation in project viva: an epigenome-wide association study. Environ Health Perspect 125(8):087019. https://doi.org/10.1289/EHP1246
Cardenas A, Rifas-Shiman SL, Agha G, Hivert MF, Litonjua AA, DeMeo DL, Lin X, Amarasiriwardena CJ, Oken E, Gillman MW, Baccarelli AA (2017) Persistent DNA methylation changes associated with prenatal mercury exposure and cognitive performance during childhood. Sci Rep 7(1):288. https://doi.org/10.1038/s41598-017-00384-5
Cardenas A, Koestler DC, Houseman EA, Jackson BP, Kile ML, Karagas MR, Marsit CJ (2015) Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics 10(6):508–515. https://doi.org/10.1080/15592294.2015.1046026
Cardenas A, Rifas-Shiman SL, Godderis L, Duca RC, Navas-Acien A, Litonjua AA, DeMeo DL, Brennan KJ, Amarasiriwardena CJ, Hivert MF, Gillman MW, Oken E, Baccarelli AA (2017) Prenatal exposure to mercury: associations with global DNA methylation and Hydroxymethylation in cord blood and in childhood. Environ Health Perspect 125(8):087022. https://doi.org/10.1289/EHP1467
Lozano M, Yousefi P, Broberg K, Soler-Blasco R, Miyashita C, Pesce G, Kim WJ, Rahman M, Bakulski KM, Haug LS, Ikeda-Araki A, Huel G, Park J, Relton C, Vrijheid M, Rifas-Shiman S, Oken E, Dou JF, Kishi R, Gutzkow KB, Annesi-Maesano I, Won S, Hivert MF, Fallin MD, Vafeiadi M, Ballester F, Bustamante M, Llop S (2021) DNA methylation changes associated with prenatal mercury exposure: a meta-analysis of prospective cohort studies from PACE consortium. Environ Res 204(Pt B):112093. https://doi.org/10.1016/j.envres.2021.112093
Gibb HJ, Lees PS, Pinsky PF, Rooney BC (2000) Lung cancer among workers in chromium chemical production. Am J Ind Med 38(2):115–126. https://doi.org/10.1002/1097-0274(200008)38:2<115::aid-ajim1>3.0.co;2-y
Zhitkovich A, Shrager S, Messer J (2000) Reductive metabolism of Cr(VI) by cysteine leads to the formation of binary and ternary Cr – DNA adducts in the absence of oxidative DNA damage. Chem Res Toxicol 13(11):1114–1124. https://doi.org/10.1021/tx0001169
Shi H, Hudson LG, Liu KJ (2004) Oxidative stress and apoptosis in metal ion-induced carcinogenesis. Free Radic Biol Med 37(5):582–593. https://doi.org/10.1016/j.freeradbiomed.2004.03.012
Kondo K, Takahashi Y, Hirose Y, Nagao T, Tsuyuguchi M, Hashimoto M, Ochiai A, Monden Y, Tangoku A (2006) The reduced expression and aberrant methylation of p16(INK4a) in chromate workers with lung cancer. Lung Cancer 53(3):295–302. https://doi.org/10.1016/j.lungcan.2006.05.022
Schnekenburger M, Talaska G, Puga A (2007) Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation. Mol Cell Biol 27(20):7089–7101. https://doi.org/10.1128/MCB.00838-07
Vineis P, Husgafvel-Pursiainen K (2005) Air pollution and cancer: biomarker studies in human populations. Carcinogenesis 26(11):1846–1855. https://doi.org/10.1093/carcin/bgi216
Samet JM, Dominici F, Curriero FC, Coursac I, Zeger SL (2000) Fine particulate air pollution and mortality in 20 U.S. cities, 1987–1994. N Engl J Med 343(24):1742–1749. https://doi.org/10.1056/NEJM200012143432401
Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, Cantone L, Rizzo G, Hou L, Schwartz J, Bertazzi PA, Baccarelli A (2009) Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect 117(2):217–222. https://doi.org/10.1289/ehp.11898
Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J (2009) Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med 179(7):572–578. https://doi.org/10.1164/rccm.200807-1097OC
Wang C, O’Brien KM, Xu Z, Sandler DP, Taylor JA, Weinberg CR (2020) Long-term ambient fine particulate matter and DNA methylation in inflammation pathways: results from the sister study. Epigenetics 15(5):524–535. https://doi.org/10.1080/15592294.2019.1699894
Chi GC, Liu Y, MacDonald JW, Barr RG, Donohue KM, Hensley MD, Hou L, McCall CE, Reynolds LM, Siscovick DS, Kaufman JD (2016) Long-term outdoor air pollution and DNA methylation in circulating monocytes: results from the multi-ethnic study of atherosclerosis (MESA). Environ Health 15(1):119. https://doi.org/10.1186/s12940-016-0202-4
Madrigano J, Baccarelli A, Mittleman MA, Wright RO, Sparrow D, Vokonas PS, Tarantini L, Schwartz J (2011) Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect 119(7):977–982. https://doi.org/10.1289/ehp.1002773
Wu Y, Qie R, Cheng M, Zeng Y, Huang S, Guo C, Zhou Q, Li Q, Tian G, Han M, Zhang Y, Wu X, Li Y, Zhao Y, Yang X, Feng Y, Liu D, Qin P, Hu D, Hu F, Xu L, Zhang M (2021) Air pollution and DNA methylation in adults: a systematic review and meta-analysis of observational studies. Environ Pollut 284:117152. https://doi.org/10.1016/j.envpol.2021.117152
Liu X, Ye Y, Chen Y, Li X, Feng B, Cao G, Xiao J, Zeng W, Li X, Sun J, Ning D, Yang Y, Yao Z, Guo Y, Wang Q, Zhang Y, Ma W, Du Q, Zhang B, Liu T (2019) Effects of prenatal exposure to air particulate matter on the risk of preterm birth and roles of maternal and cord blood LINE-1 methylation: a birth cohort study in Guangzhou. China Environ Int 133(Pt A):105177. https://doi.org/10.1016/j.envint.2019.105177
Yang P, Gong YJ, Cao WC, Wang RX, Wang YX, Liu C, Chen YJ, Huang LL, Ai SH, Lu WQ, Zeng Q (2018) Prenatal urinary polycyclic aromatic hydrocarbon metabolites, global DNA methylation in cord blood, and birth outcomes: a cohort study in China. Environ Pollut 234:396–405. https://doi.org/10.1016/j.envpol.2017.11.082
Breton CV, Yao J, Millstein J, Gao L, Siegmund KD, Mack W, Whitfield-Maxwell L, Lurmann F, Hodis H, Avol E, Gilliland FD (2016) Prenatal air pollution exposures, DNA methyl transferase genotypes, and associations with Newborn LINE1 and Alu methylation and childhood blood pressure and carotid intima-media thickness in the Children’s health study. Environ Health Perspect 124(12):1905–1912. https://doi.org/10.1289/EHP181
Cai J, Zhao Y, Liu P, Xia B, Zhu Q, Wang X, Song Q, Kan H, Zhang Y (2017) Exposure to particulate air pollution during early pregnancy is associated with placental DNA methylation. Sci Total Environ 607–608:1103–1108. https://doi.org/10.1016/j.scitotenv.2017.07.029
Ren C, Park SK, Vokonas PS, Sparrow D, Wilker E, Baccarelli A, Suh HH, Tucker KL, Wright RO, Schwartz J (2010) Air pollution and homocysteine: more evidence that oxidative stress-related genes modify effects of particulate air pollution. Epidemiology 21(2):198–206. https://doi.org/10.1097/EDE.0b013e3181cc8bfc
Baccarelli A, Zanobetti A, Martinelli I, Grillo P, Hou L, Lanzani G, Mannucci PM, Bertazzi PA, Schwartz J (2007) Air pollution, smoking, and plasma homocysteine. Environ Health Perspect 115(2):176–181. https://doi.org/10.1289/ehp.9517
Eze IC, Jeong A, Schaffner E, Rezwan FI, Ghantous A, Foraster M, Vienneau D, Kronenberg F, Herceg Z, Vineis P, Brink M, Wunderli JM, Schindler C, Cajochen C, Roosli M, Holloway JW, Imboden M, Probst-Hensch N (2020) Genome-wide DNA methylation in peripheral blood and Long-term exposure to source-specific transportation noise and air pollution: the SAPALDIA study. Environ Health Perspect 128(6):67003. https://doi.org/10.1289/EHP6174
Gondalia R, Baldassari A, Holliday KM, Justice AE, Mendez-Giraldez R, Stewart JD, Liao D, Yanosky JD, Brennan KJM, Engel SM, Jordahl KM, Kennedy E, Ward-Caviness CK, Wolf K, Waldenberger M, Cyrys J, Peters A, Bhatti P, Horvath S, Assimes TL, Pankow JS, Demerath EW, Guan W, Fornage M, Bressler J, North KE, Conneely KN, Li Y, Hou L, Baccarelli AA, Whitsel EA (2019) Methylome-wide association study provides evidence of particulate matter air pollution-associated DNA methylation. Environ Int 132:104723. https://doi.org/10.1016/j.envint.2019.03.071
Panni T, Mehta AJ, Schwartz JD, Baccarelli AA, Just AC, Wolf K, Wahl S, Cyrys J, Kunze S, Strauch K, Waldenberger M, Peters A (2016) Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the normative aging study. Environ Health Perspect 124(7):983–990. https://doi.org/10.1289/ehp.1509966
Sayols-Baixeras S, Fernandez-Sanles A, Prats-Uribe A, Subirana I, Plusquin M, Kunzli N, Marrugat J, Basagana X, Elosua R (2019) Association between long-term air pollution exposure and DNA methylation: the REGICOR study. Environ Res 176:108550. https://doi.org/10.1016/j.envres.2019.108550
Li H, Chen R, Cai J, Cui X, Huang N, Kan H (2018) Short-term exposure to fine particulate air pollution and genome-wide DNA methylation: a randomized, double-blind, crossover trial. Environ Int 120:130–136. https://doi.org/10.1016/j.envint.2018.07.041
Gruzieva O, Xu CJ, Yousefi P, Relton C, Merid SK, Breton CV, Gao L, Volk HE, Feinberg JI, Ladd-Acosta C, Bakulski K, Auffray C, Lemonnier N, Plusquin M, Ghantous A, Herceg Z, Nawrot TS, Pizzi C, Richiardi L, Rusconi F, Vineis P, Kogevinas M, Felix JF, Duijts L, den Dekker HT, Jaddoe VWV, Ruiz JL, Bustamante M, Anto JM, Sunyer J, Vrijheid M, Gutzkow KB, Grazuleviciene R, Hernandez-Ferrer C, Annesi-Maesano I, Lepeule J, Bousquet J, Bergstrom A, Kull I, Soderhall C, Kere J, Gehring U, Brunekreef B, Just AC, Wright RJ, Peng C, Gold DR, Kloog I, DeMeo DL, Pershagen G, Koppelman GH, London SJ, Baccarelli AA, Melen E (2019) Prenatal particulate air pollution and DNA methylation in Newborns: an epigenome-wide meta-analysis. Environ Health Perspect 127(5):57012. https://doi.org/10.1289/EHP4522
Gruzieva O, Xu CJ, Breton CV, Annesi-Maesano I, Anto JM, Auffray C, Ballereau S, Bellander T, Bousquet J, Bustamante M, Charles MA, de Kluizenaar Y, den Dekker HT, Duijts L, Felix JF, Gehring U, Guxens M, Jaddoe VV, Jankipersadsing SA, Merid SK, Kere J, Kumar A, Lemonnier N, Lepeule J, Nystad W, Page CM, Panasevich S, Postma D, Slama R, Sunyer J, Soderhall C, Yao J, London SJ, Pershagen G, Koppelman GH, Melen E (2017) Epigenome-wide meta-analysis of methylation in children related to prenatal NO2 air pollution exposure. Environ Health Perspect 125(1):104–110. https://doi.org/10.1289/EHP36
Ferrari L, Carugno M, Bollati V (2019) Particulate matter exposure shapes DNA methylation through the lifespan. Clin Epigenetics 11(1):129. https://doi.org/10.1186/s13148-019-0726-x
Isaevska E, Moccia C, Asta F, Cibella F, Gagliardi L, Ronfani L, Rusconi F, Stazi MA, Richiardi L (2021) Exposure to ambient air pollution in the first 1000 days of life and alterations in the DNA methylome and telomere length in children: a systematic review. Environ Res 193:110504. https://doi.org/10.1016/j.envres.2020.110504
Rider CF, Carlsten C (2019) Air pollution and DNA methylation: effects of exposure in humans. Clin Epigenetics 11(1):131. https://doi.org/10.1186/s13148-019-0713-2
White AJ, Kresovich JK, Keller JP, Xu Z, Kaufman JD, Weinberg CR, Taylor JA, Sandler DP (2019) Air pollution, particulate matter composition and methylation-based biologic age. Environ Int 132:105071. https://doi.org/10.1016/j.envint.2019.105071
Robinson BW, Lake RA (2005) Advances in malignant mesothelioma. N Engl J Med 353(15):1591–1603. https://doi.org/10.1056/NEJMra050152
Sugarbaker DJ, Richards WG, Gordon GJ, Dong L, De Rienzo A, Maulik G, Glickman JN, Chirieac LR, Hartman ML, Taillon BE, Du L, Bouffard P, Kingsmore SF, Miller NA, Farmer AD, Jensen RV, Gullans SR, Bueno R (2008) Transcriptome sequencing of malignant pleural mesothelioma tumors. Proc Natl Acad Sci U S A 105(9):3521–3526. https://doi.org/10.1073/pnas.0712399105
Christensen BC, Godleski JJ, Marsit CJ, Houseman EA, Lopez-Fagundo CY, Longacker JL, Bueno R, Sugarbaker DJ, Nelson HH, Kelsey KT (2008) Asbestos exposure predicts cell cycle control gene promoter methylation in pleural mesothelioma. Carcinogenesis 29(8):1555–1559. https://doi.org/10.1093/carcin/bgn059
He B, Lee AY, Dadfarmay S, You L, Xu Z, Reguart N, Mazieres J, Mikami I, McCormick F, Jablons DM (2005) Secreted frizzled-related protein 4 is silenced by hypermethylation and induces apoptosis in beta-catenin-deficient human mesothelioma cells. Cancer Res 65(3):743–748
Hirao T, Bueno R, Chen CJ, Gordon GJ, Heilig E, Kelsey KT (2002) Alterations of the p16(INK4) locus in human malignant mesothelial tumors. Carcinogenesis 23(7):1127–1130. https://doi.org/10.1093/carcin/23.7.1127
Lee AY, He B, You L, Dadfarmay S, Xu Z, Mazieres J, Mikami I, McCormick F, Jablons DM (2004) Expression of the secreted frizzled-related protein gene family is downregulated in human mesothelioma. Oncogene 23(39):6672–6676. https://doi.org/10.1038/sj.onc.1207881
Ohta Y, Shridhar V, Kalemkerian GP, Bright RK, Watanabe Y, Pass HI (1999) Thrombospondin-1 expression and clinical implications in malignant pleural mesothelioma. Cancer 85(12):2570–2576
Tsou JA, Galler JS, Wali A, Ye W, Siegmund KD, Groshen S, Laird PW, Turla S, Koss MN, Pass HI, Laird-Offringa IA (2007) DNA methylation profile of 28 potential marker loci in malignant mesothelioma. Lung Cancer 58(2):220–230. https://doi.org/10.1016/j.lungcan.2007.06.015
Christensen BC, Houseman EA, Godleski JJ, Marsit CJ, Longacker JL, Roelofs CR, Karagas MR, Wrensch MR, Yeh RF, Nelson HH, Wiemels JL, Zheng S, Wiencke JK, Bueno R, Sugarbaker DJ, Kelsey KT (2009) Epigenetic profiles distinguish pleural mesothelioma from normal pleura and predict lung asbestos burden and clinical outcome. Cancer Res 69(1):227–234. https://doi.org/10.1158/0008-5472.CAN-08-2586
Kettunen E, Hernandez-Vargas H, Cros MP, Durand G, Le Calvez-Kelm F, Stuopelyte K, Jarmalaite S, Salmenkivi K, Anttila S, Wolff H, Herceg Z, Husgafvel-Pursiainen K (2017) Asbestos-associated genome-wide DNA methylation changes in lung cancer. Int J Cancer 141(10):2014–2029. https://doi.org/10.1002/ijc.30897
Guarrera S, Viberti C, Cugliari G, Allione A, Casalone E, Betti M, Ferrante D, Aspesi A, Casadio C, Grosso F, Libener R, Piccolini E, Mirabelli D, Dianzani I, Magnani C, Matullo G (2019) Peripheral blood DNA methylation as potential biomarker of malignant pleural mesothelioma in Asbestos-exposed subjects. J Thorac Oncol 14(3):527–539. https://doi.org/10.1016/j.jtho.2018.10.163
Cugliari G, Allione A, Russo A, Catalano C, Casalone E, Guarrera S, Grosso F, Ferrante D, Sculco M, La Vecchia M, Pirazzini C, Libener R, Mirabelli D, Magnani C, Dianzani I, Matullo G (2021) New DNA methylation signals for malignant pleural mesothelioma risk assessment. Cancers (Basel) 13(11). https://doi.org/10.3390/cancers13112636
Oner D, Ghosh M, Moisse M, Duca RC, Coorens R, Vanoirbeek JAJ, Lambrechts D, Godderis L, Hoet PHM (2018) Global and gene-specific DNA methylation effects of different asbestos fibres on human bronchial epithelial cells. Environ Int 115:301–311. https://doi.org/10.1016/j.envint.2018.03.031
Oner D, Ghosh M, Coorens R, Bove H, Moisse M, Lambrechts D, Ameloot M, Godderis L, Hoet PHM (2020) Induction and recovery of CpG site specific methylation changes in human bronchial cells after long-term exposure to carbon nanotubes and asbestos. Environ Int 137:105530. https://doi.org/10.1016/j.envint.2020.105530
Wang J, Tian X, Zhang J, Tan L, Ouyang N, Jia B, Chen C, Ge C, Li J (2021) Postchronic single-walled carbon nanotube exposure causes irreversible malignant transformation of human bronchial epithelial cells through DNA methylation changes. ACS Nano 15(4):7094–7104. https://doi.org/10.1021/acsnano.1c00239
Feinberg AP, Cui H, Ohlsson R (2002) DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Semin Cancer Biol 12(5):389–398. https://doi.org/10.1016/s1044-579x(02)00059-7
Feinberg AP (2000) DNA methylation, genomic imprinting and cancer. Curr Top Microbiol Immunol 249:87–99. https://doi.org/10.1007/978-3-642-59696-4_6
Feinberg AP (2004) The epigenetics of cancer etiology. Semin Cancer Biol 14(6):427–432. https://doi.org/10.1016/j.semcancer.2004.06.005
Tilghman SM (1999) The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell 96(2):185–193. https://doi.org/10.1016/s0092-8674(00)80559-0
Wolffe AP (2000) Transcriptional control: imprinting insulation. Curr Biol 10(12):R463–R465. https://doi.org/10.1016/s0960-9822(00)00534-0
Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses JJ, Reik W, Feil R (1998) Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development 125(12):2273–2282
Young LE, Sinclair KD, Wilmut I (1998) Large offspring syndrome in cattle and sheep. Rev Reprod 3(3):155–163. https://doi.org/10.1530/ror.0.0030155
Cox GF, Burger J, Lip V, Mau UA, Sperling K, Wu BL, Horsthemke B (2002) Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet 71(1):162–164. https://doi.org/10.1086/341096
DeBaun MR, Niemitz EL, Feinberg AP (2003) Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet 72(1):156–160. https://doi.org/10.1086/346031
Jones PA, Liang G (2009) Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 10(11):805–811. https://doi.org/10.1038/nrg2651
Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, Leslie RD, Deloukas P, Spector TD (2010) Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res 20(4):434–439. https://doi.org/10.1101/gr.103101.109
Masuda T, Shinoara H, Kondo M (1975) Reactions of hydroxyl radicals with nucleic acid bases and the related compounds in gamma-irradiated aqueous solution. J Radiat Res 16(3):153–161. https://doi.org/10.1269/jrr.16.153
Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC (2004) Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res 32(14):4100–4108. https://doi.org/10.1093/nar/gkh739
Valinluck V, Sowers LC (2007) Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res 67(3):946–950. https://doi.org/10.1158/0008-5472.CAN-06-3123
Henderson JP, Byun J, Takeshita J, Heinecke JW (2003) Phagocytes produce 5-chlorouracil and 5-bromouracil, two mutagenic products of myeloperoxidase, in human inflammatory tissue. J Biol Chem 278(26):23522–23528. https://doi.org/10.1074/jbc.M303928200
Sun FL, Elgin SC (1999) Putting boundaries on silence. Cell 99(5):459–462. https://doi.org/10.1016/s0092-8674(00)81534-2
Turker MS (2002) Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene 21(35):5388–5393. https://doi.org/10.1038/sj.onc.1205599
Turker MS, Bestor TH (1997) Formation of methylation patterns in the mammalian genome. Mutat Res 386(2):119–130. https://doi.org/10.1016/s1383-5742(96)00048-8
Muse ME, Titus AJ, Salas LA, Wilkins OM, Mullen C, Gregory KJ, Schneider SS, Crisi GM, Jawale RM, Otis CN, Christensen BC, Arcaro KF (2020) Enrichment of CpG island shore region hypermethylation in epigenetic breast field cancerization. Epigenetics 15(10):1093–1106. https://doi.org/10.1080/15592294.2020.1747748
Schwartz DA, Freedman JH, Linney EA (2004) Environmental genomics: a key to understanding biology, pathophysiology and disease. Hum Mol Genet 13 Spec No 2:R217–224. https://doi.org/10.1093/hmg/ddh228
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Christensen, B.C., Everson, T.M., Marsit, C.J., Kelsey, K.T. (2022). Influence of Environmental Factors on the Epigenome. In: Michels, K.B. (eds) Epigenetic Epidemiology. Springer, Cham. https://doi.org/10.1007/978-3-030-94475-9_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-94475-9_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-94474-2
Online ISBN: 978-3-030-94475-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)