Abstract
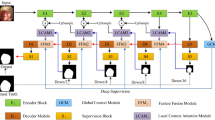
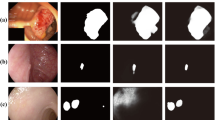
Automatic polyp segmentation is considered indispensable in modern polyp screening systems. It can help the clinicians accurately locate polyp areas for further diagnosis or surgeries. Benefit from the advancement of deep learning techniques, various neural networks are developed for handling the polyp segmentation problem. However, most of these methods neither aggregate multi-scale or multi-receptive-field features nor consider the area-boundary constraints. To address these issues, we propose a novel selective feature aggregation network with the area and boundary constraints. The network contains a shared encoder and two mutually constrained decoders for predicting polyp areas and boundaries, respectively. Feature aggregation is achieved by (1) introducing three up-concatenations between encoder and decoders and (2) embedding Selective Kernel Modules into convolutional layers which can adaptively extract features from different size of kernels. We call these two operations the Selective Feature Aggregation. Furthermore, a new boundary-sensitive loss function is proposed to take into account the dependency between the area and boundary branch, thus two branches can be reciprocally influenced and enable more accurate area predictions. We evaluate our method on the EndoScene dataset and achieve the state-of-the-art results with a Dice of 83.08% and a Accuracy of 96.68%.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
American cancer society: Key statistics for colorectal cancer. http://www.cancer.org/cancer/colonandrectumcancer/detailedguide/colorectal-cancer-keystatistics/. Accessed 18 Mar 2019
Akbari, M., et al.: Polyp segmentation in colonoscopy images using fully convolutional network. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 69–72. IEEE (2018)
Brandao, P., et al.: Fully convolutional neural networks for polyp segmentation in colonoscopy. In: Medical Imaging 2017: Computer-Aided Diagnosis, vol. 10134, p. 101340F. International Society for Optics and Photonics (2017)
Li, X., et al.: Selective kernel networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 510–519 (2019)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Milletari, F., Navab, N., Ahmadi, S.A.: V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE (2016)
Murugesan, B., et al.: Joint shape learning and segmentation for medical images using a minimalistic deep network. arXiv preprint arXiv:1901.08824 (2019)
Murugesan, B., et al.: Psi-Net: shape and boundary aware joint multi-task deep network for medical image segmentation. arXiv preprint arXiv:1902.04099 (2019)
Oda, H., et al.: BESNet: boundary-enhanced segmentation of cells in histopathological images. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 228–236. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_26
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Vázquez, et al.: A benchmark for endoluminal scene segmentation of colonoscopy images. J. Healthc. Eng. 2017 (2017)
Wickstrøm, K., Kampffmeyer, M., Jenssen, R.: Uncertainty and interpretability in convolutional neural networks for semantic segmentation of colorectal polyps. arXiv preprint arXiv:1807.10584 (2018)
Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., Liang, J.: UNet++: a nested U-Net architecture for medical image segmentation. In: Stoyanov, D., et al. (eds.) DLMIA/ML-CDS -2018. LNCS, vol. 11045, pp. 3–11. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00889-5_1
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Fang, Y., Chen, C., Yuan, Y., Tong, Ky. (2019). Selective Feature Aggregation Network with Area-Boundary Constraints for Polyp Segmentation. In: Shen, D., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science(), vol 11764. Springer, Cham. https://doi.org/10.1007/978-3-030-32239-7_34
Download citation
DOI: https://doi.org/10.1007/978-3-030-32239-7_34
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32238-0
Online ISBN: 978-3-030-32239-7
eBook Packages: Computer ScienceComputer Science (R0)