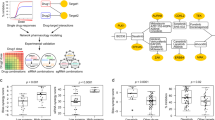
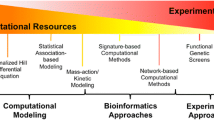
Abstract
The widespread development of resistance to cancer monotherapies has prompted the need to identify combinatorial treatment approaches that circumvent drug resistance and achieve more durable clinical benefit. However, given the vast space of possible combinations of existing drugs, the inaccessibility of drug screens to candidate targets with no available drugs, and the significant heterogeneity of cancers, exhaustive experimental testing of combination treatments remains highly impractical. There is thus an urgent need to develop computational approaches that complement experimental efforts and aid the identification and prioritization of effective drug combinations. Here, we provide a practical guide to SynDISCO, a computational framework that leverages mechanistic ODE modeling to predict and prioritize synergistic combination treatments directed at signaling networks. We demonstrate the key steps of SynDISCO and its application to the EGFR-MET signaling network in triple negative breast cancer as an illustrative example. SynDISCO is, however, a network- and cancer-independent framework, and given a suitable ODE model of the network of interest, it could be leveraged to discover cancer-specific combination treatments.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Labrie M, Brugge JS, Mills GB, Zervantonakis IK (2022) Therapy resistance: opportunities created by adaptive responses to targeted therapies in cancer. Nat Rev Cancer 22(6):323–339
Cremers CG, Nguyen LK (2019) Network rewiring, adaptive resistance and combating strategies in breast cancer. Cancer Drug Resist 2(4):1106–1126
Jaaks P et al (2022) Effective drug combinations in breast, colon and pancreatic cancer cells. Nature 603(7899):166–173
Kolch W, Halasz M, Granovskaya M, Kholodenko BN (2015) The dynamic control of signal transduction networks in cancer cells. Nat Rev Cancer 15(9):515–527
Romano D et al (2014) Protein interaction switches coordinate Raf-1 and MST2/Hippo signalling. Nat Cell Biol 16(7):673–684
Roskoski R Jr (2021) Properties of FDA-approved small molecule protein kinase inhibitors: a 2021 update. Pharmacol Res 165:105463
Ghomlaghi M, Hart A, Hoang N, Shin S, Nguyen LK (2021) Feedback, crosstalk and competition: ingredients for emergent non-linear behaviour in the PI3K/mTOR signalling network. Int J Mol Sci 22(13):6944
Nguyen LK, Kholodenko BN (2016) Feedback regulation in cell signalling: lessons for cancer therapeutics. Semin Cell Dev Biol 50:85–94
Montagud A et al (2022) Patient-specific Boolean models of signalling networks guide personalised treatments. elife 11:e72626
Frank TD, Cavadas MAS, Nguyen LK, Cheong A (2016) Non-linear dynamics in transcriptional regulation: biological logic gates. In: Carballido-Landeira J, Escribano B (eds) Nonlinear dynamics in biological systems. Springer International Publishing, Cham, pp 43–62
Toni T, Stumpf MPH (2009) Simulation-based model selection for dynamical systems in systems and population biology. Bioinformatics 26(1):104–110
Aldridge BB, Saez-Rodriguez J, Muhlich JL, Sorger PK, Lauffenburger DA (2009) Fuzzy logic analysis of kinase pathway crosstalk in TNF/EGF/insulin-induced signaling. PLoS Comput Biol 5(4):e1000340
Shin S-Y, Müller A-K, Verma N, Lev S, Nguyen LK (2018) Systems modelling of the EGFR-PYK2-c-Met interaction network predicts and prioritizes synergistic drug combinations for triple-negative breast cancer. PLoS Comput Biol 14(6):e1006192
Nguyen LK et al (2013) A dynamic model of the hypoxia-inducible factor 1α (HIF-1α) network. J Cell Sci 126(Pt 6):1454–1463
Bouhaddou M et al (2018) A mechanistic pan-cancer pathway model informed by multi-omics data interprets stochastic cell fate responses to drugs and mitogens. PLoS Comput Biol 14(3):e1005985
Ghomlaghi M, Yang G, Shin S-Y, James DE, Nguyen LK (2021) Dynamic modelling of the PI3K/MTOR signalling network uncovers biphasic dependence of mTORC1 activity on the mTORC2 subunit SIN1. PLoS Comput Biol 17(9):e1008513
Shin SY, Nguyen LK (2017) Dissecting cell-fate determination through integrated mathematical modeling of the ERK/MAPK signaling pathway. Methods Mol Biol 1487:409–432
Tyson JJ et al (2011) Dynamic modelling of oestrogen signalling and cell fate in breast cancer cells. Nat Rev Cancer 11(7):523–532
Chowdhury S, Sarkar RR (2015) Comparison of human cell signaling pathway databases—evolution, drawbacks and challenges. Database 2015:bau126
Gillespie M et al (2021) The reactome pathway knowledgebase 2022. Nucleic Acids Res 50(D1):D687–D692
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Licata L et al (2020) SIGNOR 2.0, the SIGnaling Network Open Resource 2.0: 2019 update. Nucleic Acids Res 48(D1):D504–d510
Martens M et al (2021) WikiPathways: connecting communities. Nucleic Acids Res 49(D1):D613–d621
Malik-Sheriff RS et al (2020) BioModels-15 years of sharing computational models in life science. Nucleic Acids Res 48(D1):D407–d415
Villaverde AF, Pathirana D, Fröhlich F, Hasenauer J, Banga JR (2022) A protocol for dynamic model calibration. Brief Bioinform 23(1):bbab387
Wenthur CJ, Gentry PR, Mathews TP, Lindsley CW (2014) Drugs for allosteric sites on receptors. Annu Rev Pharmacol Toxicol 54:165–184
Sebaugh JL (2011) Guidelines for accurate EC50/IC50 estimation. Pharm Stat 10(2):128–134
Li X et al (2016) β-elemene sensitizes hepatocellular carcinoma cells to oxaliplatin by preventing oxaliplatin-induced degradation of copper transporter 1. Sci Rep 6:21010
Liu F, Shang Y, Chen S-z (2014) Chloroquine potentiates the anti-cancer effect of lidamycin on non-small cell lung cancer cells in vitro. Acta Pharmacol Sin 35(5):645–652
Keith CT, Borisy AA, Stockwell BR (2005) Multicomponent therapeutics for networked systems. Nat Rev Drug Discov 4(1):71–78
Zhao W et al (2014) A new bliss Independence model to analyze drug combination data. J Biomol Screen 19(5):817–821
Chou TC (2010) Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70(2):440–446
Berenbaum MC (1989) What is synergy? Pharmacol Rev 41(2):93–141
Yadav B, Wennerberg K, Aittokallio T, Tang J (2015) Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput Struct Biotechnol J 13:504–513
Ianevski A, Giri AK, Aittokallio T (2020) SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res 48(W1):W488–W493
Arab-Bafrani Z, Shahbazi-Gahrouei D, Abbasian M, Fesharaki M (2016) Multiple MTS assay as the alternative method to determine survival fraction of the irradiated HT-29 colon cancer cells. J Med Signals Sens 6(2):112–116
Sellés Vidal L, Kelly CL, Mordaka PM, Heap JT (2018) Review of NAD(P)H-dependent oxidoreductases: properties, engineering and application. Biochim Biophys Acta Proteins Proteom 1866(2):327–347
Franken NAP, Rodermond HM, Stap J, Haveman J, van Bree C (2006) Clonogenic assay of cells in vitro. Nat Protoc 1(5):2315–2319
Hanson KM, Finkelstein JN (2019) An accessible and high-throughput strategy of continuously monitoring apoptosis by fluorescent detection of caspase activation. Anal Biochem 564-565:96–101
Kho D et al (2015) Application of xCELLigence RTCA biosensor technology for revealing the profile and window of drug responsiveness in real time. Biosensors 5(2):199–222
Nakai K, Hung MC, Yamaguchi H (2016) A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res 6(8):1609–1623
Verma N et al (2017) Targeting of PYK2 synergizes with EGFR antagonists in basal-like TNBC and circumvents HER3-associated resistance via the NEDD4–NDRG1 axis. Cancer Res 77(1):86–99
Ho-Yen CM, Jones JL, Kermorgant S (2015) The clinical and functional significance of c-Met in breast cancer: a review. Breast Cancer Res 17(1):52
Linklater ES et al (2016) Targeting MET and EGFR crosstalk signaling in triple-negative breast cancers. Oncotarget 7(43):69903–69915
Qi J et al (2011) Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res 71(3):1081–1091
Shin S-Y et al (2014) The switching role of β-adrenergic receptor signalling in cell survival or death decision of cardiomyocytes. Nat Commun 5(1):5777
Shin D et al (2014) The hidden switches underlying RORα-mediated circuits that critically regulate uncontrolled cell proliferation. J Mol Cell Biol 6(4):338–348
Masuda H et al (2012) Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat 136(2):331–345
Zhu Z et al (2021) Eucannabinolide, a novel sesquiterpene lactone, suppresses the growth, metastasis and BCSCS-like traits of TNBC via inactivation of STAT3. Neoplasia 23(1):36–48
Chaudhary SS et al (2020) Chapter 11 – c-Met as a potential therapeutic target in triple negative breast cancer. In: Gupta SP (ed) Cancer-leading proteases. Academic, San Diego, California, United States, pp 295–326
Pedersen MW, Pedersen N, Ottesen LH, Poulsen HS (2005) Differential response to gefitinib of cells expressing normal EGFR and the mutant EGFRvIII. Br J Cancer 93(8):915–923
Han S et al (2009) Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design. J Biol Chem 284(19):13193–13201
Dussault I, Bellon SF (2009) From concept to reality: the long road to c-Met and RON receptor tyrosine kinase inhibitors for the treatment of cancer. Anti Cancer Agents Med Chem 9(2):221–229
Allen JV et al (2011) The discovery of benzanilides as c-Met receptor tyrosine kinase inhibitors by a directed screening approach. Bioorg Med Chem Lett 21(18):5224–5229
Schust J, Sperl B, Hollis A, Mayer TU, Berg T (2006) Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol 13(11):1235–1242
Shin S-Y et al (2021) Integrative modelling of signalling network dynamics identifies cell type-selective therapeutic strategies for FGFR4-driven cancers. bioRxiv:2021.2011.2003.467180
Frohlich F et al (2018) Efficient parameter estimation enables the prediction of drug response using a mechanistic pan-cancer pathway model. Cell Syst 7(6):567–579 e566
Loewe S (1953) The problem of synergism and antagonism of combined drugs. Arzneimittelforschung 3(6):285–290
Zheng S et al (2022) SynergyFinder plus: toward better interpretation and annotation of drug combination screening datasets. J Pharmacokinet Pharmacodyn 20(3):587–596. https://doi.org/10.1016/j.gpb.2022.01.004
Di Veroli GY et al (2016) Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 32(18):2866–2868
Chou TC (2006) Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 58(3):621–681
Acknowledgments
This work was supported by a Victorian Cancer Agency Mid-Career Research Fellowship (MCRF18026), an Investigator Initiated Research Scheme grant from National Breast Cancer Foundation (IIRS-20-094), and a Metcalf Venture Grant by Cancer Council Victoria, Australia, awarded to L.K.N.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Appendices
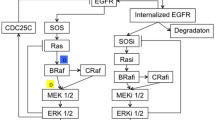
Appendix A. Ordinary Differential Equations of the EGFR-MET Model
The reaction rates are given in Appendix B.
Left-hand sides | Right-hand sides | Initial conditions (nM) |
|---|---|---|
d[pEGFR]/dt | v1 − v2 | 0.109 |
d[EGFRub]/dt | v3 − v4 | 6.940 |
d[PYK2m]/dt | v5 − v6 | 0.622 |
d[PYK2]/dt | v7 − v8 − v9 + v10 | 9.299 |
d[pPYK2]/dt | v9 − v10 | 2.510 |
d[pSTAT3]/dt | v11 − v12 | 1.178 |
d[cMETm]/dt | v13 − v14 | 0.023 |
d[cMET]/dt | v15 − v16 − v17 + v18 | 4.672 |
d[pcMET]/dt | v17 − v18 | 0.502 |
d[pCbl]/dt | v19 − v20 | 10.476 |
d[aPTP]/dt | v21 − v22 | 0.494 |
d[pERK]/dt | v23 − v24 | 0.669 |
d[STAT3uStattic]/dt | v25 | 0.00 |
Appendix B. Reactions and Reaction Rates of the EGFR-MET Network Model
Reaction | Reaction rates | |
|---|---|---|
v1 | EGFR → pEGFR | kc1 * (EGF/(1 + Gefitinib/Ki1) + caEGF) * EGFR/(Km1 + EGFR) |
v2 | pEGFR → EGFR | (Vmax2 + kc2 * aPTP) * pEGFR/(Km2 + pEGFR) |
v3 | EGFR → EGFRub | (Vmax3 + kc3 * pCbl) * EGFR/(Km3 + EGFR) * Ki3a/(Ki3a + PYK2tot/(1 + PF396/Ki3b)) |
v4 | EGFRub → EGFR | Vmax4 * EGFRub/(Km4 + EGFRub) |
v5 | ∅ → PYK2m | Vs5 + Vmax5 * pSTAT3/(Km5 + pSTAT3) |
v6 | PYK2m → ∅ | kdeg6 * PYK2m |
v7 | PYK2m → PYK2 | Vmax7 * PYK2m/(Km7 + PYK2m) |
v8 | PYK2 → ∅ | kdeg8 * PYK2 |
v9 | PYK2 → pPYK2 | (kc9a * pEGFR + kc9b * pcMET/(1 + EMD/Ki9)) * PYK2/(Km9 + PYK2) |
v10 | pPYK2 → PYK2 | (Vmax10 + kc10 * aPTP) * pPYK2/(Km10 + pPYK2) |
v11 | STAT3 → pSTAT3 | kc11 * (pPYK2/(1 + PF396/Ki3b)) * STAT3/(Km11 + STAT3) |
v12 | pSTAT3 → STAT3 | (Vmax12 + kc12 * aPTP) * pSTAT3/(Km12 + pSTAT3) |
v13 | ∅ → cMETm | Vs13 + Vmax13 * pSTAT3/(Km13 + pSTAT3) |
v14 | cMETm→ ∅ | kdeg14 * cMETm |
v15 | cMETm → cMET | Vmax15 * cMETm/(Km15 + cMETm) |
v16 | cMET → ∅ | (kdeg16 + kc16 * pCbl) * cMET/(Km16 + cMET) |
v17 | cMET → pcMET | (kc17 * HGF + caHGF) * cMET/(Km17 + cMET) |
v18 | pcMET → cMET | Vmax18 * pcMET/(Km18 + pcMET) |
v19 | Cbl → pCbl | kc19 * pEGFR * Cbl/(Km19 + Cbl) |
v20 | pCbl → Cbl | (Vmax20 + kc20 * aPTP) * pCbl/(Km20 + pCbl) |
v21 | PTP → aPTP | kc21 * pEGFR * PTP/(Km21 + PTP) |
v22 | aPTP → PTP | Vmax22 * aPTP/(Km22 + aPTP) |
v23 | ERK → pERK | (kc23a * pcMET/(1 + EMD/Ki23) + kc23b * pEGFR) * ERK/(Km23 + ERK) |
v24 | pERK → ERK | Vmax24 * pERK/(Km24 + pERK) |
v25 | STAT3 + Stattic ↔ STAT3uStattic | ka25 * STAT3 * Stattic − kd25 * STAT3uStattic |
Appendix C. Best-Fitted Parameter Values Used for Simulations
Parameter | Value | Unit | Parameter | Value | Unit |
|---|---|---|---|---|---|
kc1 | 413.0475 | min−1 | Km17 | 9.817479 | nM |
Km1 | 248.8857 | nM | Vmax18 | 0.060674 | nM min−1 |
Ki1 | 1.0000 | μM | Km18 | 9.954054 | nM |
kc2 | 1406.048 | min−1 | kdeg16 | 24.49063 | min−1 |
Km2 | 3.801894 | nM | kc16 | 1.174898 | min−1 |
Vmax3 | 0.000104 | nM min−1 | Km16 | 528.4453 | nM |
Km3 | 2.285599 | nM | kc19 | 52.72299 | min−1 |
Ki3a | 0.08356 | nM | Km19 | 13.30454 | nM |
Ki3b | 1.0000 | nM | kc20 | 35.64511 | min−1 |
Vmax4 | 11.11732 | nM min−1 | Km20 | 24.32204 | nM |
kc3 | 10.78947 | min−1 | kc21 | 0.003972 | min−1 |
Km4 | 90.78205 | nM | Km21 | 52.72299 | nM |
Vs5 | 26.54606 | nM min−1 | Vmax22 | 0.034914 | nM min−1 |
Vmax5 | 34.04082 | nM min−1 | Km22 | 46.45153 | nM |
Km5 | 4.74242 | nM | Vmax2 | 112.2018 | nM min−1 |
kdeg6 | 53.57967 | min−1 | Vmax12 | 7.638358 | nM min−1 |
Vmax7 | 3.349654 | nM min−1 | Vmax20 | 0.048306 | nM min−1 |
Km7 | 3.334264 | nM | kc23a | 7.03E + 09 | min−1 |
kc9a | 0.463447 | min−1 | kc23b | 8.43E + 08 | min−1 |
kc9b | 0.988553 | min−1 | Km23 | 2.831392 | nM |
Km9 | 34.91403 | nM | Vmax24 | 4.4E + 09 | nM min−1 |
Vmax10 | 0.530884 | nM min−1 | Km24 | 0.156675 | nM |
Km10 | 9.141132 | nM | kc10 | 0.006109 | min−1 |
kdeg8 | 0.056624 | min−1 | Ki9 | 1.65577 | nM |
kc11 | 0.321366 | min−1 | Ki23 | 13.48963 | nM |
Km11 | 20.6063 | nM | ka25 | 127.3503 | μM−1 min−1 |
kc12 | 0.00029 | min−1 | kd25 | 11.74898 | min−1 |
Km12 | 11.58777 | nM | caEGF | 0.089125 | nM |
Vs13 | 0.093756 | nM min−1 | caHGF | 0.009036 | nM |
Vmax13 | 0.354813 | nM min−1 | EGFRtot | 398.1072 | nM |
Km13 | 38.72576 | nM | STAT3tot | 144.2115 | nM |
kdeg14 | 4.560369 | min−1 | Cbltot | 174.9847 | nM |
Vmax15 | 91.41132 | nM min−1 | PTPtot | 296.4831 | nM |
Km15 | 6.456542 | nM | ERKtot | 166.7247 | nM |
kc17 | 0.000811 | min−1 |
Rights and permissions
Copyright information
© 2023 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Shin, SY., Nguyen, L.K. (2023). SynDISCO: A Mechanistic Modeling-Based Framework for Predictive Prioritization of Synergistic Drug Combinations Targeting Cell Signalling Networks. In: Nguyen, L.K. (eds) Computational Modeling of Signaling Networks. Methods in Molecular Biology, vol 2634. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3008-2_17
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3008-2_17
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3007-5
Online ISBN: 978-1-0716-3008-2
eBook Packages: Springer Protocols