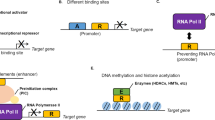
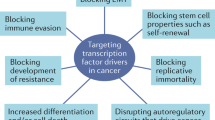
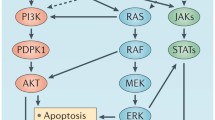
Abstract
Deregulation of transcription factors is critical to hallmarks of cancer. Genetic mutations, gene fusions, amplifications or deletions, epigenetic alternations, and aberrant post-transcriptional modification of transcription factors are involved in the regulation of various stages of carcinogenesis, including cancer initiation, progression, and metastasis. Thus, targeting the dysfunctional transcription factors may lead to new cancer therapeutic strategies. However, transcription factors are conventionally considered as “undruggable.” Here, we summarize the recent progresses in understanding the regulation of transcription factors in cancers and strategies to target transcription factors and co-factors for preclinical and clinical drug development, particularly focusing on c-Myc, YAP/TAZ, and β-catenin due to their significance and interplays in cancer.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Latchman DS (1997) Transcription factors: an overview. Int J Biochem Cell Biol 29(12):1305–1312
Darnell JE (2002) Transcription factors as targets for cancer therapy. Nat Rev Cancer 2(10):740–749
Bushweller JH (2019) Targeting transcription factors in cancer—from undruggable to reality. Nat Rev Cancer 19(11):611–624
Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K et al (2010) An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140(5):744–752
Pobbati AV, Hong W (2020) A combat with the YAP/TAZ-TEAD oncoproteins for cancer therapy. Theranostics 10(8):3622
Gill MK, Christova T, Zhang YY, Gregorieff A, Zhang L, Narimatsu M et al (2018) A feed forward loop enforces YAP/TAZ signaling during tumorigenesis. Nat Commun 9(1):1–13
Meyer N, Penn LZ (2008) Reflecting on 25 years with MYC. Nat Rev Cancer 8(12):976–990
Kalkat M, De Melo J, Hickman KA, Lourenco C, Redel C, Resetca D et al (2017) MYC deregulation in primary human cancers. Gene 8(6):151
Clevers H (2006) Wnt/β-catenin signaling in development and disease. Cell 127(3):469–480
Moon RT, Kohn AD, De Ferrari GV, Kaykas A (2004) WNT and β-catenin signalling: diseases and therapies. Nat Rev Genet 5(9):691–701
Wang T, Zheng L, Wang Q, Hu YW (2018) Emerging roles and mechanisms of FOXC2 in cancer. Clin Chim Acta 479:84–93
Alasiri G, Fan LYN, Zona S, Goldsbrough IG, Ke HL, Auner HW, Lam EWF (2018) ER stress and cancer: the FOXO forkhead transcription factor link. Mol Cell Endocrinol 462:67–81
Li Y, Zhang Y, Yao Z, Li S, Yin Z, Xu M (2016) Forkhead box Q1: a key player in the pathogenesis of tumors. Int J Oncol 49(1):51–58
Han B, Bhowmick N, Qu Y, Chung S, Giuliano AE, Cui X (2017) FOXC1: an emerging marker and therapeutic target for cancer. Oncogene 36(28):3957–3963
Nestal de Moraes G, Carneiro LDT, Maia RC, Lam EWF, Sharrocks AD (2019) FOXK2 transcription factor and its emerging roles in cancer. Cancer 11(3):393
Gartel AL (2017) FOXM1 in cancer: interactions and vulnerabilities. Cancer Res 77(12):3135–3139
Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W (2017) FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci 13(7):815
Lee H, Jeong AJ, Ye SK (2019) Highlighted STAT3 as a potential drug target for cancer therapy. BMB Rep 52(7):415
Kaltschmidt C, Banz-Jansen C, Benhidjeb T, Beshay M, Förster C, Greiner J et al (2019) A role for NF-κB in organ specific cancer and cancer stem cells. Cancer 11(5):655
Otálora-Otálora BA, Henríquez B, López-Kleine L, Rojas A (2019) RUNX family: Oncogenes or tumor suppressors. Oncol Rep 42(1):3–19
Khachigian LM (2018) The Yin and Yang of YY 1 in tumor growth and suppression. Int J Cancer 143(3):460–465
Atsaves V, Leventaki V, Rassidakis GZ, Claret FX (2019) AP-1 transcription factors as regulators of immune responses in cancer. Cancer 11(7):1037
Muller PA, Vousden KH (2013) p53 mutations in cancer. Nat Cell Biol 15(1):2–8
Whibley C, Pharoah PD, Hollstein M (2009) p53 polymorphisms: cancer implications. Nat Rev Cancer 9(2):95–107
de la Vega MR, Chapman E, Zhang DD (2018) NRF2 and the Hallmarks of Cancer. Cancer Cell 34(1):21–43
Arkin MR, Tang Y, Wells JA (2014) Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol 21(9):1102–1114
Arkin MR, Wells JA (2004) Small-molecule inhibitors of protein–protein interactions: progressing towards the dream. Nat Rev Drug Discov 3(4):301–317
Silvian LF, Friedman JE, Strauch K, Cachero TG, Day ES, Qian F et al (2011) Small molecule inhibition of the TNF family cytokine CD40 ligand through a subunit fracture mechanism. ACS Chem Biol 6(6):636–647
Illendula A, Gilmour J, Grembecka J, Tirumala VSS, Boulton A, Kuntimaddi A et al (2016) Small molecule inhibitor of CBFβ-RUNX binding for RUNX transcription factor driven cancers. EBioMedicine 8:117–131
Li T, Kang G, Wang T, Huang H (2018) Tumor angiogenesis and anti-angiogenic gene therapy for cancer. Oncol Lett 16(1):687–702
Zaimy MA, Saffarzadeh N, Mohammadi A, Pourghadamyari H, Izadi P, Sarli A et al (2017) New methods in the diagnosis of cancer and gene therapy of cancer based on nanoparticles. Cancer Gene Ther 24(6):233–243
Hiemer SE, Zhang L, Kartha VK, Packer TS, Almershed M, Noonan V et al (2015) A YAP/TAZ-regulated molecular signature is associated with oral squamous cell carcinoma. Mol Cancer Res 13(6):957–968
Bisso A, Filipuzzi M, Figueroa GPG, Brumana G, Biagioni F, Doni M et al (2019) Cooperation between MYC and β-catenin in liver tumorigenesis requires. Yap/Taz bioRxiv:819631
Wu Q, Li J, Sun S, Chen X, Zhang H, Li B, Sun S (2017) YAP/TAZ-mediated activation of serine metabolism and methylation regulation is critical for LKB1-deficient breast cancer progression. Biosci Rep 37(5):1–6
Wang C, Jeong K, Jiang H, Guo W, Gu C, Lu Y, Liang J (2016) YAP/TAZ regulates the insulin signaling via IRS1/2 in endometrial cancer. Am J Cancer Res 6(5):996
Horie M, Saito A, Ohshima M, Suzuki HI, Nagase T (2016) YAP and TAZ modulate cell phenotype in a subset of small cell lung cancer. Cancer Sci 107(12):1755–1766
Chanvorachote P, Sriratanasak N, Nonpanya N (2020) C-myc contributes to malignancy of lung cancer: a potential anticancer drug target. Anticancer Res 40(2):609–618
Elbadawy M, Usui T, Yamawaki H, Sasaki K (2019) Emerging roles of C-Myc in Cancer stem cell-related signaling and resistance to cancer chemotherapy: a potential therapeutic target against colorectal cancer. Int J Mol Sci 20(9):2340
Miyoshi K, Hennighausen L (2003) β-Catenin: a transforming actor on many stages. Breast Cancer Research 5(2):63
Kypta RM, Waxman J (2012) Wnt/β-catenin signalling in prostate cancer. Nature Rev Urol 9(8):418
Elian FA, Yan E, Walter MA (2018) FOXC1, the new player in the cancer sandbox. Oncotarget 9(8):8165
Nandi D, Cheema PS, Jaiswal N, Nag A (2018) FoxM1: repurposing an oncogene as a biomarker. In: Seminars in cancer biology. Academic Press, pp 74–84
Laissue P (2019) The forkhead-box family of transcription factors: key molecular players in colorectal cancer pathogenesis. Mol Cancer 18(1):1–13
Grossi V, Fasano C, Celestini V, Lepore Signorile M, Sanese P, Simone C (2019) Chasing the FOXO3: Insights into Its New Mitochondrial Lair in Colorectal Cancer Landscape. Cancer 11(3):414
Zhang J, Niu Y, Huang C (2017) Role of FoxM1 in the progression and epithelial to mesenchymal transition of gastrointestinal Cancer. Recent Patents Anti-Cancer Drug Dis 12(3):247–259
Brenner O, Levanon D, Negreanu V, Golubkov O, Fainaru O, Woolf E, Groner Y (2004) Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc Natl Acad Sci 101(45):16016–16021
Araki K, Osaki M, Nagahama Y, Hiramatsu T, Nakamura H, Ohgi S, Ito H (2005) Expression of RUNX3 protein in human lung adenocarcinoma: implications for tumor progression and prognosis. Cancer Sci 96(4):227–231
Blyth K, Cameron ER, Neil JC (2005) The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer 5(5):376–387
Endo T, Ohta K, Kobayashi T (2008) Expression and function of Cbfa-1/Runx2 in thyroid papillary carcinoma cells. J Clin Endocrinol Metabol 93(6):2409–2412
Chuang LSH, Ito K, Ito Y (2017) Roles of RUNX in solid tumors. In: RUNX Proteins in Development and Cancer. Springer, Singapore, pp 299–320
Sun J, Li B, Jia Z, Zhang A, Wang G, Chen Z et al (2018) RUNX3 inhibits glioma survival and invasion via suppression of the β-catenin/TCF-4 signaling pathway. J Neuro-Oncol 140(1):15–26
Sweeney K, Cameron ER, Blyth K (2020) Complex Interplay between the RUNX Transcription Factors and Wnt/β-Catenin Pathway in Cancer: A Tango in the Night. Mol Cell 43(2):188
Tang C, Zhu G (2019) Classic and novel signaling pathways involved in cancer: targeting the NF-ΚB and syk signaling pathways. Curr Stem Cell Res Ther 14(3):219–225
Ferraiuolo M, Pulito C, Finch-Edmondson M, Korita E, Maidecchi A, Donzelli S et al (2018) Agave negatively regulates YAP and TAZ transcriptionally and post-translationally in osteosarcoma cell lines. Cancer Lett 433:18–32
Koh CM, Sabò A, Guccione E (2016) Targeting MYC in cancer therapy: RNA processing offers new opportunities. BioEssays 38(3):266–275
Collins S, Groudine M (1982) Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature 298(5875):679–681
Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F et al (1983) Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 305(5931):245–248
Nau MM, Brooks BJ, Battey J, Sausville E, Gazdar AF, Kirsch IR et al (1985) L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature 318(6041):69–73
Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C (2013) Emerging landscape of oncogenic signatures across human cancers. Nat Genet 45(10):1127–1133
Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B et al (2013) Pan-cancer patterns of somatic copy number alteration. Nat Genet 45(10):1134–1140
Wierstra I, Alves J (2008) The c-myc promoter: still MysterY and challenge. Adv Cancer Res 99:113–333
Cowling VH, Turner SA, Cole MD (2014) Burkitt’s lymphoma-associated c-Myc mutations converge on a dramatically altered target gene response and implicate Nol5a/Nop56 in oncogenesis. Oncogene 33(27):3519–3527
Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V et al (2016) Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet 48(4):398
Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I (1993) Point mutations in the c–Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet 5(1):56–61
Bahram F, von der Lehr N, Cetinkaya C, Larsson LG (2000) c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood J Am Soc Hematol 95(6):2104–2110
Symonds G, Hartshorn A, Kennewell A, O'Mara MA, Bruskin A, Bishop JM (1989) Transformation of murine myelomonocytic cells by myc: point mutations in v-myc contribute synergistically to transforming potential. Oncogene 4(3):285–294
Chakraborty AA, Scuoppo C, Dey S, Thomas LR, Lorey SL, Lowe SW, Tansey WP (2015) A common functional consequence of tumor-derived mutations within c-MYC. Oncogene 34(18):2406–2409
Nair SK, Burley SK (2006) Structural aspects of interactions within the Myc/Max/Mad network. In: The Myc/Max/Mad Transcription Factor Network. Springer, Berlin, Heidelberg, pp 123–143
Channavajhala P, Seldin DC (2002) Functional interaction of protein kinase CK2 and c-Myc in lymphomagenesis. Oncogene 21(34):5280–5288
Hann, S. R. (2006). Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. In Seminars in cancer biology(Vol. 16, No. 4, pp. 288-302). Academic Press
Vervoorts J, Lüscher-Firzlaff J, Lüscher B (2006) The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem 281(46):34725–34729
Sears RC (2004) The life cycle of C-myc: from synthesis to degradation. Cell Cycle 3(9):1131–1135
Benassi B, Fanciulli M, Fiorentino F, Porrello A, Chiorino G, Loda M et al (2006) c-Myc phosphorylation is required for cellular response to oxidative stress. Mol Cell 21(4):509–519
Henriksson M, Bakardjiev A, Klein G, Lüscher B (1993) Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene 8(12):3199–3209
Lutterbach B, Hann SR (1994) Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol 14(8):5510–5522
Lutterbach B, Hann SR (1999) c-Myc transactivation domain-associated kinases: Questionable role for map kinases in c-Myc phosphorylation. J Cell Biochem 72(4):483–491
Noguchi K, Kitanaka C, Yamana H, Kokubu A, Mochizuki T, Kuchino Y (1999) Regulation of c-Myc through phosphorylation at Ser-62 and Ser-71 by c-Jun N-terminal kinase. J Biol Chem 274(46):32580–32587
Sears R, Leone G, DeGregori J, Nevins JR (1999) Ras enhances Myc protein stability. Mol Cell 3(2):169–179
Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR (2000) Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14(19):2501–2514
Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP (2003) Skp2 regulates Myc protein stability and activity. Mol Cell 11(5):1177–1188
Von Der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C et al (2003) The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell 11(5):1189–1200
Molinari E, Gilman M, Natesan S (1999) Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J 18(22):6439–6447
Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, Krek W (2001) Skp2 is oncogenic and overexpressed in human cancers. Proc Natl Acad Sci 98(9):5043–5048
Zeng L, Zhou MM (2002) Bromodomain: an acetyl-lysine binding domain. FEBS Lett 513(1):124–128
Vervoorts J, Lüscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K et al (2003) Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep 4(5):484–490
Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ et al (2004) The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol 24(24):10826–10834
Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E et al (2005) Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol 25(23):10220–10234
Piccolo S, Dupont S, Cordenonsi M (2014) The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev 94(4):1287–1312
Pan D (2010) The hippo signaling pathway in development and cancer. Dev Cell 19(4):491–505
Meng Z, Moroishi T, Guan KL (2016) Mechanisms of Hippo pathway regulation. Genes Dev 30(1):1–17
Gill MK, Christova T, Zhang YY, Gregorieff A, Zhang L, Narimatsu M et al A feed forward loop enforces YAP/TAZ signaling during tumorigenesis. Nat Commun 9(1):1–13
Johnson R, Halder G (2014) The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov 13(1):63–79
Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C et al (2011) The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147(4):759–772
Liu JY, Li YH, Lin HX, Liao YJ, Mai SJ, Liu ZW et al (2013) Overexpression of YAP 1 contributes to progressive features and poor prognosis of human urothelial carcinoma of the bladder. BMC Cancer 13(1):349
Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J et al (2006) Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125(7):1253–1267
Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D et al (2011) Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 144(5):782–795
St John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG et al (1999) Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet 21(2):182–186
Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X (2010) Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci 101(5):1279–1285
Strnadel J, Choi S, Fujimura K, Wang H, Zhang W, Wyse M et al (2017) eIF5A-PEAK1 signaling regulates YAP1/TAZ protein expression and pancreatic cancer cell growth. Cancer Res 77(8):1997–2007
Felley-Bosco E, Stahel R (2014) Hippo/YAP pathway for targeted therapy. Translat Lung Cancer Res 3(2):75
Chan P, Han X, Zheng B, DeRan M, Yu J, Jarugumilli GK et al (2016) Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat Chem Biol 12(4):282
Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J et al (2016) Tead and AP1 coordinate transcription and motility. Cell Rep 14(5):1169–1180
Kim MK, Jang JW, Bae SC (2018) DNA binding partners of YAP/TAZ. BMB Rep 51(3):126
Ji J, Xu R, Zhang X, Han M, Xu Y, Wei Y et al (2018) Actin like-6A promotes glioma progression through stabilization of transcriptional regulators YAP/TAZ. Cell Death Dis 9(5):1–16
Zhang Z, Du J, Wang S, Shao L, Jin K, Li F et al (2019) OTUB2 promotes cancer metastasis via hippo-independent activation of YAP and TAZ. Mol Cell 73(1):7–21
Yao F, Zhou Z, Kim J, Hang Q, Xiao Z, Ton BN et al (2018) SKP2-and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity. Nat Commun 9(1):1–16
Polakis P (2000) Wnt signaling and cancer. Genes Dev 14(15):1837–1851
Miyoshi K, Hennighausen L (2003) β-Catenin: a transforming actor on many stages. Breast Cancer Research. 5(2):63
Chan E, Gat U, McNiff JM, Fuchs E (1999) A common human skin tumour is caused by activating mutations in β-catenin. Nat Genet 21(4):410–413
Wan X, Liu J, Lu JF, Tzelepi V, Yang J, Starbuck MW et al (2012) Activation of β-catenin signaling in androgen receptor–negative prostate Cancer cells. Clin Cancer Res 18(3):726–736
Khurana N, Sikka SC (2019) Interplay between SOX9, Wnt/β-catenin and androgen receptor signaling in castration-resistant prostate cancer. Int J Mol Sci 20(9):2066
Aberle H, Bauer A, Stappert J, Kispert A, Kemler R (1997) β-catenin is a target for the ubiquitin–proteasome pathway. EMBO J 16(13):3797–3804
Tetsu O, McCormick F (1999) β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398(6726):422–426
Nussinov R, Tsai CJ, Jang H, Korcsmáros T, Csermely P (2016) Oncogenic KRAS signaling and YAP1/β-catenin: Similar cell cycle control in tumor initiation. In: Seminars in cell & developmental biology. Academic Press, p 85
Deng F, Peng L, Li Z, Tan G, Liang E, Chen S et al (2018) YAP triggers the Wnt/β-catenin signalling pathway and promotes enterocyte self-renewal, regeneration and tumorigenesis after DSS-induced injury. Cell Death Dis 9(2):1–16
Liu H, Du S, Lei T, Wang H, He X, Tong R, Wang Y (2018) Multifaceted regulation and functions of YAP/TAZ in tumors. Oncol Rep 40(1):16–28
Li Q, Sun Y, Jarugumilli GK, Liu S, Dang K, Cotton JL et al (2020) Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription. Cell Stem Cell 26(5):675–692
Sun J, Wang X, Tang B, Liu H, Zhang M, Wang Y et al (2018) A tightly controlled Src-YAP signaling axis determines therapeutic response to dasatinib in renal cell carcinoma. Theranostics 8(12):3256
Moroishi T, Hansen CG, Guan KL (2015) The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer 15(2):73–79
Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S et al (2014) Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol 16(4):357–366
Wang Z, Wu Y, Wang H, Zhang Y, Mei L, Fang X et al (2014) Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc Natl Acad Sci 111(1):E89–E98
Guo J, Wu Y, Yang L, Du J, Gong K, Chen W et al (2017) Repression of YAP by NCTD disrupts NSCLC progression. Oncotarget 8(2):2307
Bao Y, Nakagawa K, Yang Z, Ikeda M, Withanage K, Ishigami-Yuasa M et al (2011) A cell-based assay to screen stimulators of the Hippo pathway reveals the inhibitory effect of dobutamine on the YAP-dependent gene transcription. J Biochemi 150(2):199–208
Zheng HX, Wu LN, Xiao H, Du Q, Liang JF (2014) Inhibitory effects of dobutamine on human gastric adenocarcinoma. World J Gastroenterol: WJG 20(45):17092
Lee SB, Gong YD, Park YI, Dong MS (2013) 2, 3, 6-Trisubstituted quinoxaline derivative, a small molecule inhibitor of the Wnt/beta-catenin signaling pathway, suppresses cell proliferation and enhances radiosensitivity in A549/Wnt2 cells. Biochem Biophys Res Commun 431(4):746–752
Lee SB, Park YI, Dong MS, Gong YD (2010) Identification of 2, 3, 6-trisubstituted quinoxaline derivatives as a Wnt2/β-catenin pathway inhibitor in non-small-cell lung cancer cell lines. Bioorg Med Chem Lett 20(19):5900–5904
Park S, Choi J (2010) Inhibition of β-catenin/Tcf signaling by flavonoids. J Cell Biochem 110(6):1376–1385
Amado NG, Predes D, Moreno MM, Carvalho IO, Mendes FA, Abreu JG (2014) Flavonoids and Wnt/β-catenin signaling: potential role in colorectal cancer therapies. Int J Mol Sci 15(7):12094–12106
Li X, Bai B, Liu L, Ma P, Kong L, Yan J et al (2015) Novel β-carbolines against colorectal cancer cell growth via inhibition of Wnt/β-catenin signaling. Cell Death Dis 1(1):1–9
Kong L, Mao B, Zhu H, Li Y (2015) Novel β-carbolines inhibit Wnt/β-catenin signaling.
Castell A, Yan Q, Fawkner K, Hydbring P, Zhang F, Verschut V et al (2018) A selective high affinity MYC-binding compound inhibits MYC: MAX interaction and MYC-dependent tumor cell proliferation. Sci Rep 8(1):1–17
Yap JL, Wang H, Hu A, Chauhan J, Jung KY, Gharavi RB et al (2013) Pharmacophore identification of c-Myc inhibitor 10074-G5. Bioorg Med Chem Lett 23(1):370–374
Chauhan J, Wang H, Yap JL, Sabato PE, Hu A, Prochownik EV, Fletcher S (2014) Discovery of Methyl 4′-Methyl-5-(7-nitrobenzo [c][1, 2, 5] oxadiazol-4-yl)-[1, 1′-biphenyl]-3-carboxylate, an Improved Small-Molecule Inhibitor of c-Myc–Max Dimerization. ChemMedChem 9(10):2274–2285
Hart JR, Garner AL, Yu J, Ito Y, Sun M, Ueno L et al (2014) Inhibitor of MYC identified in a Kröhnke pyridine library. Proc Natl Acad Sci 111(34):12556–12561
Jeong KC, Kim KT, Seo HH, Shin SP, Ahn KO, Ji MJ et al (2014) Intravesical instillation of c-MYC inhibitor KSI-3716 suppresses orthotopic bladder tumor growth. J Urol 191(2):510–518
Choi SH, Mahankali M, Lee SJ, Hull M, Petrassi HM, Chatterjee AK et al (2017) Targeted disruption of Myc–Max oncoprotein complex by a small molecule. ACS Chem Biol 12(11):2715–2719
Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X et al (2014) A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25(2):166–180
Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA et al (2012) Genetic and pharmacological disruption of the TEAD–YAP complex suppresses the oncogenic activity of YAP. Genes Dev 26(12):1300–1305
Tschaharganeh DF, Chen X, Latzko P, Malz M, Gaida MM, Felix K et al (2013) Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology 144(7):1530–1542
Brodowska K, Al-Moujahed A, Marmalidou A, Zu Horste MM, Cichy J, Miller JW, Vavvas DG (2014) The clinically used photosensitizer Verteporfin (VP) inhibits YAP-TEAD and human retinoblastoma cell growth in vitro without light activation. Exp Eye Res 124:67–73
Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E et al (2015) The Hippo effector YAP promotes resistance to RAF-and MEK-targeted cancer therapies. Nat Genet 47(3):250–256
Kim MH, Kim J, Hong H, Lee SH, Lee JK, Jung E, Kim J (2016) Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J 35(5):462–478
Fisher ML, Grun D, Adhikary G, Xu W, Eckert RL (2017) Inhibition of YAP function overcomes BRAF inhibitor resistance in melanoma cancer stem cells. Oncotarget 8(66):110257
Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M et al (2004) A small molecule inhibitor of β-catenin/cyclic AMP response element-binding protein transcription. Proc Natl Acad Sci 101(34):12682–12687
Manegold P, Lai KK, Wu Y, Teo JL, Lenz HJ, Genyk YS et al (2018) Differentiation therapy targeting the β-catenin/CBP interaction in pancreatic cancer. Cancer 10(4):95
Dietrich L, Rathmer B, Ewan K, Bange T, Heinrichs S, Dale TC et al (2017) Cell permeable stapled peptide inhibitor of Wnt signaling that targets β-catenin protein-protein interactions. Cell Chem Biol 24(8):958–968
Gonsalves FC, Klein K, Carson BB, Katz S, Ekas LA, Evans S et al (2011) An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc Natl Acad Sci 108(15):5954–5963
Sukhdeo K, Mani M, Zhang Y, Dutta J, Yasui H, Rooney MD et al (2007) Targeting the β-catenin/TCF transcriptional complex in the treatment of multiple myeloma. Proc Natl Acad Sci 104(18):7516–7521
Li X, Pu J, Jiang S, Su J, Kong L, Mao B et al (2013) Henryin, an ent-kaurane diterpenoid, inhibits Wnt signaling through interference with β-catenin/TCF4 interaction in colorectal cancer cells. PLoS One 8(7): 1–10
Wang Z, Zhang M, Wang J, Ji H (2019) Optimization of Peptidomimetics as Selective Inhibitors for the β-Catenin/T-Cell Factor Protein–Protein Interaction. J Med Chem 62(7):3617–3635
Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S et al (1992) Peptoids: a modular approach to drug discovery. Proc Natl Acad Sci 89(20):9367–9371
Schneider JA, Craven TW, Kasper AC, Yun C, Haugbro M, Briggs EM et al (2018) Design of Peptoid-peptide Macrocycles to Inhibit the β-catenin TCF Interaction in Prostate Cancer. Nat Commun 9(1):1–10
Wang H, Teriete P, Hu A, Raveendra-Panickar D, Pendelton K, Lazo JS et al (2015) Direct inhibition of c-Myc-Max heterodimers by celastrol and celastrol-inspired triterpenoids. Oncotarget 6(32):32380
Jung KY, Wang H, Teriete P, Yap JL, Chen L, Lanning ME et al (2015) Perturbation of the c-Myc–max protein–protein interaction via synthetic α-helix mimetics. J Med Chem 58(7):3002–3024
Lu JJ, Meng LH, Shankavaram UT, Zhu CH, Tong LJ, Chen G et al (2010) Dihydroartemisinin accelerates c-MYC oncoprotein degradation and induces apoptosis in c-MYC-overexpressing tumor cells. Biochem Pharmacol 80(1):22–30
Waaler J, Machon O, Tumova L, Dinh H, Korinek V, Wilson SR et al (2012) A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res 72(11):2822–2832
Hwang SY, Deng X, Byun S, Lee C, Lee SJ, Suh H et al (2016) Direct targeting of β-catenin by a small molecule stimulates proteasomal degradation and suppresses oncogenic Wnt/β-catenin signaling. Cell Rep 16(1):28–36
Yang W, Li Y, Ai Y, Obianom ON, Guo D, Yang H et al (2019) Pyrazole-4-Carboxamide (YW2065): A Therapeutic Candidate for Colorectal Cancer via Dual Activities of Wnt/β-Catenin Signaling Inhibition and AMP-Activated Protein Kinase (AMPK) Activation. J Med Chem 62(24):11151–11164
Balaji KC, Koul H, Mitra S, Maramag C, Reddy P, Menon M et al (1997) Antiproliferative effects of c-myc antisense oligonucleotide in prostate cancer cells: a novel therapy in prostate cancer. Urology 50(6):1007–1015
Iversen PL, Arora V, Acker AJ, Mason DH, Devi GR (2003) Efficacy of antisense morpholino oligomer targeted to c-myc in prostate cancer xenograft murine model and a Phase I safety study in humans. Clin Cancer Res 9(7):2510–2519
Sekhon HS, London CA, Sekhon M, Iversen PL, Devi GR (2008) c-MYC antisense phosphosphorodiamidate morpholino oligomer inhibits lung metastasis in a murine tumor model. Lung Cancer 60(3):347–354
Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res 34(19):5402–5415
Ohnmacht SA, Neidle S (2014) Small-molecule quadruplex-targeted drug discovery. Bioorg Med Chem Lett 24(12):2602–2612
Chatterjee J, Mierke DF, Kessler H (2008) Conformational preference and potential templates of N-methylated cyclic pentaalanine peptides. Chem Eur J 14(5):1508–1517
Seenisamy J, Bashyam S, Gokhale V, Vankayalapati H, Sun D, Siddiqui-Jain A et al (2005) Design and synthesis of an expanded porphyrin that has selectivity for the c-MYC G-quadruplex structure. J Am Chem Soc 127(9):2944–2959
Calabrese DR, Chen X, Leon EC, Gaikwad SM, Phyo Z, Hewitt WM et al (2018) Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat Commun 9(1):1–15
Hu MH, Wang YQ, Yu ZY, Hu LN, Ou TM, Chen SB et al (2018) Discovery of a new four-leaf clover-like ligand as a potent c-MYC transcription inhibitor specifically targeting the promoter G-quadruplex. J Med Chem 61(6):2447–2459
Ganesh S, Shui X, Craig KP, Park J, Wang W, Brown BD, Abrams MT (2018) RNAi-mediated β-catenin inhibition promotes T cell infiltration and antitumor activity in combination with immune checkpoint blockade. Mol Ther 26(11):2567–2579
Acknowledgments
The authors would like to thank Melanoma Research Alliance (MRA)-Samuel M. Fisher Memorial Established Investigator award and NIH R01s (5R01DK107651 and 1R01CA238270-01) for the support of our work. The figures are created with BioRender.com.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Tao, Z., Wu, X. (2023). Targeting Transcription Factors in Cancer: From “Undruggable” to “Druggable”. In: Song, Q., Tao, Z. (eds) Transcription Factor Regulatory Networks. Methods in Molecular Biology, vol 2594. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2815-7_9
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2815-7_9
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2814-0
Online ISBN: 978-1-0716-2815-7
eBook Packages: Springer Protocols