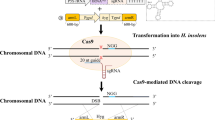
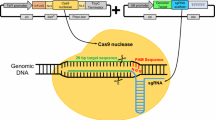
Abstract
Clostridium cellulolyticum is a model mesophilic, cellulolytic bacterium, with the potential to produce biofuels from lignocellulose. However, the natural cellulose utilization efficiency is quite low and, therefore, metabolically engineered strains with increased efficiency can decrease both the overall cost and time required for biofuel production. Traditional genetic tools are inefficient, expensive, and time-consuming, but recent developments in the use of CRISPR-Cas genetic editing systems have greatly expanded our ability to reprogram cells. Here we describe an established protocol enabling one-step versatile genome editing in C. cellulolyticum. It integrates Cas9 nickase (Cas9n) which introduces a single nick that triggers repair via homologous recombination (SNHR) to edit genomic loci with high efficiency and accuracy. This one-step editing is achieved by transforming an all-in-one vector to coexpress Cas9n and a single guide RNA (gRNA) and carries a user-defined homologous donor template to promote SNHR at a desired target site. Additionally, this system has high specificity and allows for various types of genomic editing, including markerless insertions, deletions, substitutions, and even multiplex editing.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Marraffini LA, Sontheimer EJ (2010) CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11:181
Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S (2015) Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81:2506–2514
Cobb RE, Wang Y, Zhao H (2014) High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol 4:723–728
Tong Y, Charusanti P, Zhang L, Weber T, Lee SY (2015) CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol 4:1020–1029
Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154:1370–1379
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918
Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31:833
Li J-F, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233
DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41:4336–4343
Cho SW, Kim S, Kim JM, Kim J-S (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Xu T, Li Y, Shi Z, Hemme CL, Li Y, Zhu Y, Van Nostrand JD, He Z, Zhou J (2015) Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl Environ Microbiol 81:4423–4431
Xu T, Li Y, Van Nostrand JD, He Z, Zhou J (2014) Cas9-based tools for targeted genome editing and transcriptional control. Appl Environ Microbiol 80:1544–1552
Makarova KS, Aravind L, Wolf YI, Koonin EV (2011) Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct 6:38
Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF (2011) Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol 9:467
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602
Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V (2011) The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 39:9275–9282
Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109:E2579–E2586
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM (2013) Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10:1116
Bhaya D, Davison M, Barrangou R (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45:273–297
van der Ploeg JR (2009) Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155:1966–1976
Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V (2013) crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol 10:841–851
Sorek R, Lawrence CM, Wiedenheft B (2013) CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82:237–266
Shuman S, Glickman MS (2007) Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol 5:852
Smith GR (2001) Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu Rev Genet 35:243–274
Desvaux M (2005) Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia. FEMS Microbiol Rev 29:741–764
Li Y, Tschaplinski TJ, Engle NL, Hamilton CY, Rodriguez M, Liao JC, Schadt CW, Guss AM, Yang Y, Graham DE (2012) Combined inactivation of the Clostridium cellulolyticum lactate and malate dehydrogenase genes substantially increases ethanol yield from cellulose and switchgrass fermentations. Biotechnol Biofuels 5:2
Li Y, Xu T, Tschaplinski TJ, Engle NL, Yang Y, Graham DE, He Z, Zhou J (2014) Improvement of cellulose catabolism in Clostridium cellulolyticum by sporulation abolishment and carbon alleviation. Biotechnol Biofuels 7:25
Jennert KC, Tardif C, Young DI, Young M (2000) Gene transfer to Clostridium cellulolyticum ATCC 35319. Microbiology 146:3071–3080
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822
Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, Wile BM, Vertino PM, Stewart FJ, Bao G (2014) CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res 42:7473–7485
Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S (2014) Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32:670
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Xu, T., Tao, X., Kempher, M.L., Zhou, J. (2022). Cas9 Nickase-Based Genome Editing in Clostridium cellulolyticum. In: Reisch, C.R. (eds) Recombineering. Methods in Molecular Biology, vol 2479. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2233-9_15
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2233-9_15
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2232-2
Online ISBN: 978-1-0716-2233-9
eBook Packages: Springer Protocols