Abstract
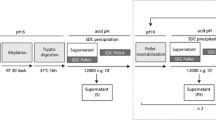
Dried blood spots (DBS) are widely used for screening molecular profiles, including enzymatic activity. However, hydrophilic proteins present in large amounts in blood inhibit detection of other proteins in DBS by liquid chromatography-mass spectrometry (LC-MS/MS) without preenrichment. Sodium carbonate precipitation (SCP) can concentrate hydrophobic proteins from DBS and effectively remove soluble hydrophilic proteins. Furthermore, SCP combination with data-independent acquisition (DIA) for quantitative LC-MS/MS enhanced the proteome analysis sensitivity and quantification limits. In this protocol, we have described in detail a simple preenrichment method using SCP and a deep proteome analysis method for LC-MS/MS data using DIA.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
McDade TW, Williams S, Snodgrass JJ (2007) What a drop can do: dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography 44(4):899–925. https://doi.org/10.1353/dem.2007.0038
Deglon J, Thomas A, Mangin P, Staub C (2012) Direct analysis of dried blood spots coupled with mass spectrometry: concepts and biomedical applications. Anal Bioanal Chem 402(8):2485–2498. https://doi.org/10.1007/s00216-011-5161-6
Li W, Tse FL (2010) Dried blood spot sampling in combination with LC-MS/MS for quantitative analysis of small molecules. Biomed Chromatogr 24(1):49–65. https://doi.org/10.1002/bmc.1367
Keevil BG (2011) The analysis of dried blood spot samples using liquid chromatography tandem mass spectrometry. Clin Biochem 44(1):110–118. https://doi.org/10.1016/j.clinbiochem.2010.06.014
Martin NJ, Bunch J, Cooper HJ (2013) Dried blood spot proteomics: surface extraction of endogenous proteins coupled with automated sample preparation and mass spectrometry analysis. J Am Soc Mass Spectrom 24(8):1242–1249. https://doi.org/10.1007/s13361-013-0658-1
Rosting C, Yu J, Cooper HJ (2018) High field asymmetric waveform ion mobility spectrometry in nontargeted bottom-up proteomics of dried blood spots. J Proteome Res 17(6):1997–2004. https://doi.org/10.1021/acs.jproteome.7b00746
Qian WJ, Kaleta DT, Petritis BO, Jiang H, Liu T, Zhang X et al (2008) Enhanced detection of low abundance human plasma proteins using a tandem IgY12-SuperMix immunoaffinity separation strategy. Mol Cell Proteomics 7(10):1963–1973. https://doi.org/10.1074/mcp.M800008-MCP200
Das S, Bosley AD, Ye X, Chan KC, Chu I, Green JE et al (2010) Comparison of strong cation exchange and SDS-PAGE fractionation for analysis of multiprotein complexes. J Proteome Res 9(12):6696–6704. https://doi.org/10.1021/pr100843x
Camerini S, Mauri P (2015) The role of protein and peptide separation before mass spectrometry analysis in clinical proteomics. J Chromatogr A 1381:1–12. https://doi.org/10.1016/j.chroma.2014.12.035
Nakajima D, Kawashima Y, Shibata H, Yasumi T, Isa M, Izawa K et al (2020) Simple and sensitive analysis for dried blood spot proteins by sodium carbonate precipitation for clinical proteomics. J Proteome Res 19(7):2821–2827. https://doi.org/10.1021/acs.jproteome.0c00271
Cao L, Clifton JG, Reutter W, Josic D (2013) Mass spectrometry-based analysis of rat liver and hepatocellular carcinoma Morris hepatoma 7777 plasma membrane proteome. Anal Chem 85(17):8112–8120. https://doi.org/10.1021/ac400774g
Kim H, Botelho SC, Park K, Kim H (2015) Use of carbonate extraction in analyzing moderately hydrophobic transmembrane proteins in the mitochondrial inner membrane. Protein Sci 24(12):2063–2069. https://doi.org/10.1002/pro.2817
Fujiki Y, Hubbard AL, Fowler S, Lazarow PB (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol 93(1):97–102. https://doi.org/10.1083/jcb.93.1.97
Ritz D, Kinzi J, Neri D, Fugmann T (2017) Data-independent acquisition of HLA class I peptidomes on the Q exactive mass spectrometer platform. Proteomics 17(19). https://doi.org/10.1002/pmic.201700177
Ting YS, Egertson JD, Bollinger JG, Searle BC, Payne SH, Noble WS et al (2017) PECAN: library-free peptide detection for data-independent acquisition tandem mass spectrometry data. Nat Methods 14(9):903–908. https://doi.org/10.1038/nmeth.4390
Tsou CC, Tsai CF, Teo GC, Chen YJ, Nesvizhskii AI (2016) Untargeted, spectral library-free analysis of data-independent acquisition proteomics data generated using Orbitrap mass spectrometers. Proteomics 16(15–16):2257–2271. https://doi.org/10.1002/pmic.201500526
Zhang Y, Bilbao A, Bruderer T, Luban J, Strambio-De-Castillia C, Lisacek F et al (2015) The use of variable Q1 isolation windows improves selectivity in LC-SWATH-MS acquisition. J Proteome Res 14(10):4359–4371. https://doi.org/10.1021/acs.jproteome.5b00543
Amodei D, Egertson J, MacLean BX, Johnson R, Merrihew GE, Keller A et al (2019) Improving precursor selectivity in data-independent acquisition using overlapping windows. J Am Soc Mass Spectrom 30(4):669–684. https://doi.org/10.1007/s13361-018-2122-8
Searle BC, Pino LK, Egertson JD, Ting YS, Lawrence RT, MacLean BX et al (2018) Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat Commun 9(1):5128. https://doi.org/10.1038/s41467-018-07454-w
Kawashima Y, Watanabe E, Umeyama T, Nakajima D, Hattori M, Honda K et al (2019) Optimization of data-independent acquisition mass spectrometry for deep and highly sensitive proteomic analysis. Int J Mol Sci 20(23):5932. https://doi.org/10.3390/ijms20235932
Holewinski RJ, Parker SJ, Matlock AD, Venkatraman V, Van Eyk JE (2016) Methods for SWATH: data independent acquisition on TripleTOF mass spectrometers. Methods Mol Biol 1410:265–279. https://doi.org/10.1007/978-1-4939-3524-6_16
Gessulat S, Schmidt T, Zolg DP, Samaras P, Schnatbaum K, Zerweck J et al (2019) Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat Methods 16(6):509–518. https://doi.org/10.1038/s41592-019-0426-7
Searle BC, Swearingen KE, Barnes CA, Schmidt T, Gessulat S, Kuster B et al (2020) Generating high quality libraries for DIA MS with empirically corrected peptide predictions. Nat Commun 11(1):1548. https://doi.org/10.1038/s41467-020-15346-1
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Nakajima, D., Ohara, O., Kawashima, Y. (2022). Data-Independent Acquisition Mass Spectrometry-Based Deep Proteome Analysis for Hydrophobic Proteins from Dried Blood Spots Enriched by Sodium Carbonate Precipitation. In: Corrales, F.J., Paradela, A., Marcilla, M. (eds) Clinical Proteomics. Methods in Molecular Biology, vol 2420. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1936-0_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1936-0_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1935-3
Online ISBN: 978-1-0716-1936-0
eBook Packages: Springer Protocols