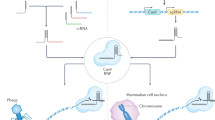
Abstract
Science selected it as the 2015 Breakthrough of the Year: Clustered Regularly Interspaced Short Palindromic Repeats, also known as CRISPR. Adaptive immunity in some bacteria and archaea allow to respond and eliminate invading genetic material; CRISPR and CRISPR-associated (Cas) genes are new molecular techniques that allow deleting, replacing or otherwise editing DNA. Using modified bacterial protein and a RNA that allows a guidance to a specific DNA sequence, CRISPR provides a striking control over several genes by not deleting the entire gene but just inactivating it by deleting few bases. CRISPR/Cas9 has been used to generate knockout cells or animals by co-expressing a gRNA specific to the gene to be targeted and the endonuclease Cas9. This novel system has been tested in diverse species, with promising potential uses in humans. Theoretically, CRISPR/Cas9 will be able to remove genetic mutations related to incurable diseases, such as HIV, and certain cancer types. This scenario draws tentative and promising conditions using CRISPR/Cas9 as preventive and therapeutic tool in medical area. As expected, several groups have begun to test the putative gene editing properties of CRISPR/Cas9 on human cells. Since sleep disorders have been linked with specific genes, in this review, we suggest areas that require further investigation and experimental and/or clinical approaches to treat sleep disturbances using CRISPR/Cas9.


Similar content being viewed by others
References
Chatterjee S, Ahituv N. Gene regulatory elements, major drivers of human disease. Annu Rev Genomics Hum Genet. 2017;18:45–63.
Petit F, Sears KE, Ahituv N. Limb development: a paradigm of gene regulation. Nat Rev Genet. 2017;18:245–58.
Yilmaz A, Grotewold E. Components and mechanisms of regulation of gene expression. Met Mol Biol. 2010;674:23–32.
Gehring NH, Wahle E, Fischer U. Deciphering the mRNP code: RNA-bound determinants of post-transcriptional gene regulation. Trends Biochem Sci. 2017;42:369–82.
Shin JH, Xu L, Wang D. Mechanism of transcription-coupled DNA modification recognition. Cell Biosci. 2017;22(7):9.
Lusk CP, King MC. The nucleus: keeping it together by keeping it apart. Curr Opin Cell Biol. 2017;44:44–50.
Baralle D, Buratti E. RNA splicing in human disease and in the clinic. Clin Sci (Lond). 2017;131:355–68.
Cheung VG, Spielman RS. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Gen. 2009;10:595–604.
Barbagallo I, Li Volti G, Galvano F, Tettamanti G, Pluchinotta FR, Bergante S, Vanella L. Diabetic human adipose tissue-derived mesenchymal stem cells fail to differentiate in functional adipocytes. Exp Biol Med. 2016;242:1079–85.
Yong LK, Lai S, Liang Z, Poteet E, Chen F, van Buren G, Fisher W, Mo Q, Chen C, Yao Q. Overexpression of Semaphorin-3E enhances pancreatic cancer cell growth and associates with poor patient survival. Oncotarget. 2016;7:87431–48.
Chen J, Li K, Pang Q, Yang C, Zhang H, Wu F, Cao H, Liu H, Wan Y, Xia W, Wang J, Dai Z, Li Y. Identification of suitable reference gene and biomarkers of serum miRNAs for osteoporosis. Sci Rep. 2016;6:36347.
American Academy of Sleep Medicine, International classification of sleep disorders: diagnostic and coding manual, 3rd edn. Darien, IL, USA.: American Academy of Sleep Medicine; 2014.
Tononi G, Cirelli C. Modulation of brain gene expression during sleep and wakefulness: a review of recent findings. Neuropsychopharmacology. 2001;25(5 Suppl):S28–35.
Cirelli C, Faraguna U, Tononi G. Changes in brain gene expression after long-term sleep deprivation. J Neurochem. 2006;98:1632–45.
Cirelli C, Pfister-Genskow M, McCarthy D, Woodbury R, Tononi G. Proteomic profiling of the rat cerebral cortex in sleep and waking. Arch Ital Biol. 2009;147:59–68.
Crocker A, Sehgal A. Genetic analysis of sleep. Genes Dev. 2010;24:1220–35.
Gehrman PR, Keenan BT, Byrne EM, Pack AI. Genetics of sleep disorders. Psyc Clin North Am. 2015;38:667–81.
Lind MJ, Gehrman PR. Genetic pathways to insomnia. Brain Sci. 2010;6:E64.
Serretti A, Benedetti F, Mandelli L, Lorenzi C, Pirovano A, Colombo C, Smeraldi E. Genetic dissection of psychopathological symptoms: insomnia in mood disorders and CLOCK gene polymorphism. Am J Med Gen. 2010;121B:35–8.
Serretti A, Gaspar-Barba E, Calati R, Cruz-Fuentes CS, Gomez-Sanchez A, Perez-Molina A, De Ronchi D. 3111T/C clock gene polymorphism is not associated with sleep disturbances in untreated depressed patients. Chronobiol Int. 2003;27:265–77.
Utge SJ, Soronen P, Loukola A, Kronholm E, Ollila HM, Pirkola S, Porkka-Heiskanen T, Partonen T, Paunio T. Systematic analysis of circadian genes in a population-based sample reveals association of TIMELESS with depression and sleep disturbance. PLoS One. 2010;5:e9259.
Brower KJ, Wojnar M, Sliwerska E, Armitage R, Burmeister M. PER3 polymorphism and insomnia severity in alcohol dependence. Sleep. 2012;35:571–7.
Li J, Huang C, Lan Y, Wang Y. A cross-sectional study on the relationships among the polymorphism of period2 gene, work stress, and insomnia. Sleep Breath. 2015;19:1399–406.
Mendlewicz J. Disruption of the circadian timing systems: molecular mechanisms in mood disorders. CNS Drugs. 2009;23(Suppl 2):15–26.
Ban HJ, Kim SC, Seo J, Kang HB, Choi JK. Genetic and metabolic characterization of insomnia. PLoS ONE. 2011;6:e18455.
Byrne EM, Gehrman PR, Medland SE, Nyholt DR, Heath AC, Madden PA, Hickie IB, Van Duijn CM, Henders AK, Montgomery GW, Martin NG, Wray NR, Chronogen Consortium. A genome-wide association study of sleep habits and insomnia. Am J Med Gen. 2013;162:439–51.
Ollila HM, Kettunen J, Pietiläinen O, Aho V, Silander K, Kronholm E, Perola M, Lahti J, Räikkönen K, Widen E, Palotie A, Eriksson JG, Partonen T, Kaprio J, Salomaa V, Raitakari O, Lehtimäki T, Sallinen M, Härmä M, Porkka-Heiskanen T, Paunio T. Genome-wide association study of sleep duration in the Finnish population. J Sleep Res. 2016;23:609–18.
Spada J, Scholz M, Kirsten H, Hensch T, Horn K, Jawinski P, Ulke C, Burkhardt R, Wirkner K, Loeffler M, Hegerl U, Sander C. Genome-wide association analysis of actigraphic sleep phenotypes in the LIFE Adult Study. J Sleep Res. 2016;25:690–701.
Subramani Y, Singh M, Wong J, Kushida CA, Malhotra A, Chung F. Understanding phenotypes of obstructive sleep apnea: applications in anesthesia, surgery, and perioperative medicine. Anesth Analg. 2017;124:179–91.
Patel N, Donahue C, Shenoy A, Patel A, El-Sherif N. Obstructive sleep apnea and arrhythmia: a systemic review. Int J Cardiol. 2017;228:967–70.
Cade BE, Chen H, Stilp AM, Gleason KJ, Sofer T, Ancoli-Israel S, Arens R, Bell GI, Below JE, Bjonnes AC, Chun S, Conomos MP, Evans DS, Johnson WC, Frazier-Wood AC, Lane JM, Larkin EK, Loredo JS, Post WS, Ramos AR, Rice K, Rotter JI, Shah NA, Stone KL, Taylor KD, Thornton TA, Tranah GJ, Wang C, Zee PC, Hanis CL, Sunyaev SR, Patel SR, Laurie CC, Zhu X, Saxena R, Lin X, Redline S. Genetic associations with obstructive sleep apnea traits in Hispanic/Latino Americans. Am J Resp Crit Care Med. 2016;194:886–97.
Huang YS, Guilleminault C, Hwang M, Cheng C, Lin CH, Li HY, Lee LA. Inflammatory cytokines in pediatric obstructive sleep apnea. Med. 2016;95:e4944.
Hirotsu C, Albuquerque RG, Nogueira H, Hachul H, Bittencourt L, Tufik S, Andersen ML. The relationship between sleep apnea, metabolic dysfunction and inflammation: the gender influence. Brain Behav Immun. 2017;59:211–8.
Wu W, Li Z, Tang T, Wu J, Liu F, Gu L. 5-HTR2A and IL-6 polymorphisms and obstructive sleep apnea-hypopnea syndrome. Biomed Rep. 2016;4:203–8.
de Lima FF, Mazzotti DR, Tufik S, Bittencourt L. The role inflammatory response genes in obstructive sleep apnea syndrome: a review. Sleep Breath. 2016;20:331–8.
Dong XS, Ma SF, Cao CW, Li J, An P, Zhao L, Liu NY, Yan H, Hu QT, Mignot E, Strohl KP, Gao ZC, Zeng C, Han F. Hypocretin (orexin) neuropeptide precursor gene, HCRT, polymorphisms in early-onset narcolepsy with cataplexy. Sleep Med. 2013;14:482–7.
Han F, Lin L, Schormair B, Pizza F, Plazzi G, Ollila HM, Nevsimalova S, Jennum P, Knudsen S, Winkelmann J, Coquillard C, Babrzadeh F, Strom TM, Wang C, Mindrinos M, Fernandez Vina M, Mignot E. HLA DQB1*06:02 negative narcolepsy with hypocretin/orexin deficiency. Sleep. 2014;37:1601–8.
Miyagawa T, Toyoda H, Hirataka A, Kanbayashi T, Imanishi A, Sagawa Y, Kotorii N, Kotorii T, Hashizume Y, Ogi K, Hiejima H, Kamei Y, Hida A, Miyamoto M, Imai M, Fujimura Y, Tamura Y, Ikegami A, Wada Y, Moriya S, Furuya H, Kato M, Omata N, Kojima H, Kashiwase K, Saji H, Khor SS, Yamasaki M, Wada Y, Ishigooka J, Kuroda K, Kume K, Chiba S, Yamada N, Okawa M, Hirata K, Uchimura N, Shimizu T, Inoue Y, Honda Y, Mishima K, Honda M, Tokunaga K. New susceptibility variants to narcolepsy identified in HLA class II region. Human Mol Gen. 2015;24:891–8.
Toyoda H, Miyagawa T, Koike A, Kanbayashi T, Imanishi A, Sagawa Y, Kotorii N, Kotorii T, Hashizume Y, Ogi K, Hiejima H, Kamei Y, Hida A, Miyamoto M, Imai M, Fujimura Y, Tamura Y, Ikegami A, Wada Y, Moriya S, Furuya H, Takeuchi M, Kirino Y, Meguro A, Remmers EF, Kawamura Y, Otowa T, Miyashita A, Kashiwase K, Khor SS, Yamasaki M, Kuwano R, Sasaki T, Ishigooka J, Kuroda K, Kume K, Chiba S, Yamada N, Okawa M, Hirata K, Mizuki N, Uchimura N, Shimizu T, Inoue Y, Honda Y, Mishima K, Honda M, Tokunaga K. A polymorphism in CCR1/CCR3 is associated with narcolepsy. Brain Behav Imm. 2015;49:148–55.
Tafti M, Lammers GJ, Dauvilliers Y, Overeem S, Mayer G, Nowak J, Pfister C, Dubois V, Eliaou JF, Eberhard HP, Liblau R, Wierzbicka A, Geisler P, Bassetti CL, Mathis J, Lecendreux M, Khatami R, Heinzer R, Haba-Rubio J, Feketeova E, Baumann CR, Kutalik Z, Tiercy JM. Narcolepsy-associated HLA class I alleles implicate cell-mediated cytotoxicity. Sleep. 2016;39:581–7.
Johansson AS, Owe-Larsson B, Hetta J, Lundkvist GB. Altered circadian clock gene expression in patients with schizophrenia. Schizophrenia Res. 2016;174:17–23.
Luo W, Ma S, Yang Y, Zhang D, Zhang Q, Liu Y, Liu Z. TFEB regulates PER3 expression via glucose-dependent effects on CLOCK/BMAL1. Int J Bioch Cell Biol. 2016;78:31–42.
Panda S. Circadian physiology of metabolism. Science. 2016;354:1008–15.
Riddle M, Mezias E, Foley D, LeSauter J, Silver R. Differential localization of PER1 and PER2 in the brain master circadian clock. Eur J Neurosci. 2016;45:1357–67.
Videnovic A, Willis GL. Circadian system—a novel diagnostic and therapeutic target in Parkinson’s disease? Mov Dis. 2016;31:260–9.
Potter GD, Skene DJ, Arendt J, Cade JE, Grant PJ, Hardie LJ. Circadian rhythm and sleep disruption: causes, metabolic consequences, and countermeasures. Endocrin Rev. 2016;37:584–608.
van Maanen A, Meijer AM, van der Heijden KB, Oort FJ. The effects of light therapy on sleep problems: a systematic review and meta-analysis. Sleep Med Rev. 2016;29:52–62.
Dijk DJ, Archer SN. PERIOD3, circadian phenotypes, and sleep homeostasis. Sleep Med Rev. 2010;14:151–60.
Nordgren A. Genes, body clocks and prevention of sleep problems. Med Health Care Phil. 2016;19:569–79.
Turek FW. Circadian clocks: not your grandfather’s clock. Science. 2016;354:992–3.
Parsons MJ, Lester KJ, Barclay NL, Archer SN, Nolan PM, Eley TC, Gregory AM. Polymorphisms in the circadian expressed genes PER3 and ARNTL2 are associated with diurnal preference and GNβ3 with sleep measures. Sleep Res. 2014;23:595–604.
Hu Y, Shmygelska A, Tran D, Eriksson N, Tung JY, Hinds DA. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat Com. 2016;7:10448.
Amin N, Allebrandt KV, van der Spek A, Müller-Myhsok B, Hek K, Teder-Laving M, Hayward C, Esko T, van Mill JG, Mbarek H, Watson NF, Melville SA, Del Greco FM, Byrne EM, Oole E, Kolcic I, Chen TH, Evans DS, Coresh J, Vogelzangs N, Karjalainen J, Willemsen G, Gharib SA, Zgaga L, Mihailov E, Stone KL, Campbell H, Brouwer RW, Demirkan A, Isaacs A, Dogas Z, Marciante KD, Campbell S, Borovecki F, Luik AI, Li M, Hottenga JJ, Huffman JE, van den Hout MC, Cummings SR, Aulchenko YS, Gehrman PR, Uitterlinden AG, Wichmann HE, Müller-Nurasyid M, Fehrmann RS, Montgomery GW, Hofman A, Kao WH, Oostra BA, Wright AF, Vink JM, Wilson JF, Pramstaller PP, Hicks AA, Polasek O, Punjabi NM, Redline S, Psaty BM, Heath AC, Merrow M, Tranah GJ, Gottlieb DJ, Boomsma DI, Martin NG, Rudan I, Tiemeier H, van Ijcken WF, Penninx BW, Metspalu A, Meitinger T, Franke L, Roenneberg T, van Duijn CM. Genetic variants in RBFOX3 are associated with sleep latency. Eur J Human Gen. 2016;24:1488–95.
Cade BE, Gottlieb DJ, Lauderdale DS, Bennett DA, Buchman AS, Buxbaum SG, De Jager PL, Evans DS, Fülöp T, Gharib SA, Johnson WC, Kim H, Larkin EK, Lee SK, Lim AS, Punjabi NM, Shin C, Stone KL, Tranah GJ, Weng J, Yaffe K, Zee PC, Patel SR, Zhu X, Redline S, Saxena R. Common variants in DRD2 are associated with sleep duration: the CARe consortium. Human Mol Gen. 2016;25:167–79.
Chang AM, Bjonnes AC, Aeschbach D. Circadian gene variants influence sleep and the sleep electroencephalogram in humans. Chronobio Int. 2016;33:561–73.
Dall’Ara I, Ghirotto S, Ingusci S, Bagarolo G, Bertolucci C, Barbujani G. Demographic history and adaptation account for clock gene diversity in humans. Heredity. 2016;117:165–72.
Truong KK, Lam MT, Grandner MA, Sassoon CS, Malhotra A. Timing matters: circadian rhythm in sepsis, obstructive lung disease, obstructive sleep apnea, and cancer. Ann Am Thor Soc. 2016;13:1144–54.
Rizzo G, Li X, Galantucci S, Filippi M, Cho YW. Brain imaging and networks in restless legs syndrome. Sleep Med. 2016;31:39–48.
Scherer JS, Combs SA, Brennan F. Sleep disorders, restless legs syndrome, and uremic pruritus: diagnosis and treatment of common symptoms in dialysis patients. Am J Kidney Dis. 2017;69:117–28.
Hennessy MD, Zak RS, Gay CL, Pullinger CR, Lee KA, Aouizerat BE. Polymorphisms of interleukin-1 Beta and interleukin-17Alpha genes are associated with restless legs syndrome. Biol Res Nurs. 2014;16:143–51.
Gan-Or Z, Zhou S, Ambalavanan A, Leblond CS, Xie P, Johnson A, Spiegelman D, Allen RP, Earley CJ, Desautels A, Montplaisir JY, Dion PA, Xiong L, Rouleau GA. Analysis of functional GLO1 variants in the BTBD9 locus and restless legs syndrome. Sleep Med. 2015;16:1151–5.
Fuh JL, Chung MY, Yao SC, Chen PK, Liao YC, Hsu CL, Wang PJ, Wang YF, Chen SP, Fann CS, Kao LS, Wang SJ. Susceptible genes of restless legs syndrome in migraine. Cephalalgia. 2016;36:1028–37.
Parish JM. Genetic and immunologic aspects of sleep and sleep disorders. Chest. 2013;143:1489–99.
Szentkirályi A, Völzke H, Hoffmann W, Winkelmann J, Berger K. Lack of association between genetic risk loci for restless legs syndrome and multimorbidity. Sleep. 2016;39:111–5.
Khan FH, Ahlberg CD, Chow CA, Shah DR, Koo BB. Iron, dopamine, genetics, and hormones in the pathophysiology of restless legs syndrome. J Neurol. 2017;264(8):1634–41.
García-Martín E, Jiménez-Jiménez FJ, Alonso-Navarro H, Martínez C, Zurdo M, Turpín-Fenoll L, Millán-Pascual J, Adeva-Bartolomé T, Cubo E, Navacerrada F, Rojo-Sebastián A, Rubio L, Ortega-Cubero S, Pastor P, Calleja M, Plaza-Nieto JF, Pilo-de-la-Fuente B, Arroyo-Solera M, García-Albea E, Agúndez JA. Heme oxygenase-1 and 2 common genetic variants and risk for restless legs syndrome. Medicine. 2015;94:e1448.
Winkelman JW, Blackwell T, Stone K, Ancoli-Israel S, Tranah GJ, Redline S, Osteoporotic Fractures in Men (MrOS) Study Research Group. Genetic associations of periodic limb movements of sleep in the elderly for the MrOS sleep study. Sleep Med. 2015;16:1360–665.
Jiménez-Jiménez FJ, García-Martí E, Alonso-Navarro H, Martínez C, Zurdo M, Turpín-Fenoll L, Millán-Pascual J, Adeva-Bartolomé T, Cubo E, Navacerrada F, Rojo-Sebastián A, Rubio L, Ortega-Cubero S, Pastor P, Calleja M, Plaza-Nieto JF, Pilo-de-la-Fuente B, Arroyo-Solera M, García-Albea E, Agúndez JA. Thr105Ile (rs11558538) polymorphism in the histamine-1-methyl-transferase (HNMT) gene and risk for restless legs syndrome. J Neural Transm. 2016;124:285–91.
Bertero A, Pawlowski M, Ortmann D, Snijders K, Yiangou L, Cardoso de Brito M, Brown S, Bernard WG, Cooper JD, Giacomelli E, Gambardella L, Hannan NR, Iyer D, Sampaziotis F, Serrano F, Zonneveld MC, Sinha S, Kotter M, Vallier L. Optimized inducible shRNA and CRISPR/Cas9 platforms for in vitro studies of human development using hPSCs. Dev. 2016;143:4405–18.
Cohen J. The birth of CRISPR Inc. Science. 2017;355:680–4.
Cohen J. CRISPR patent ruling leaves license holders scrambling. Science. 2017;355:786.
Jinek M, Chylinski K, Fonfara I, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA—guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21.
Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Gen. 2015;16:299–311.
Lin J, Zhou Y, Liu J, Chen J, Chen W, Zhao S, Wu Z, Wu N. Progress and application of CRISPR/Cas technology in biological and biomedical investigation. J Cell Biochem. 2017;118(10):3061–71.
Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotech. 2016;34:933–41.
Doerflinger M, Forsyth W, Ebert G, Pellegrini M, Herold MJ. CRISPR/Cas9-The ultimate weapon to battle infectious diseases? Cell Microbiol. 2017;19:e12693.
Koo T, Kim JS. Therapeutic applications of CRISPR RNA-guided genome editing. Brief Func Gen. 2016;16:38–45.
Oude Blenke E, Evers MJ, Mastrobattist E, van der Oost J. CRISPR-Cas9 gene editing: delivery aspects and therapeutic potential. J Control Rel. 2016;244:139–48.
Singh V, Braddick D, Dhar PK. Exploring the potential of genome editing CRISPR-Cas9 technology. Gene. 2017;599:1–18.
Strong A, Musunuru K. Genome editing in cardiovascular diseases. Nat Rev Cardiol. 2017;14:11–20.
Toth LA, Bhargava P. Animal models of sleep disorders. Comp Med. 2013;63:91–104.
Gillombardo CB, Darrah R, Dick TE. C57BL/6 J mouse apolipoprotein A2 gene is deterministic for apnea. Resp Physiol Neurobiol. 2017;235:88–94.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical standards
All data reported in this paper are from public repositories.
Funding
This work was supported by The University of California Institute for Mexico and the United States (UC MEXUS) and Consejo Nacional de Ciencia y Tecnología (CONACyT(Grant(CN-17-19) and Escuela de Medicina, Universidad Anáhuac Mayab Grant (PresInvEMR2014) given to E.M.-R.
Conflict of interest
Authors declare no conflict of interest.
Additional information
In Memoriam. Dr. René Drucker-Colín was Emeritus Researcher at National Autonomous University of Mexico as well as Head of the Office of Secretary of Science and Technology in Mexico City. Dr. Drucker-Colin passed away on Saturday, September 17th, 2017 at the age of 80. He was a world-renowned researcher in neurobiology area who received the National Science and Arts Prize (Mexico) and many other honors. Dr. Drucker-Colin’s contributions into the neurobiology of sleep–wake control have been recognized nationally and internationally. His deep commitment into social problems as well as constant enthusiasm in research will be missed by all scientific community.
Rights and permissions
About this article
Cite this article
Murillo-Rodríguez, E., Rocha, N.B., Veras, A.B. et al. The End of Snoring? Application of CRISPR/Cas9 Genome Editing for Sleep Disorders. Sleep Vigilance 2, 13–21 (2018). https://doi.org/10.1007/s41782-017-0018-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41782-017-0018-5