Abstract
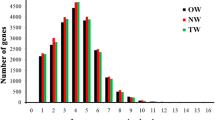
Molecular biological research into wood development and formation has been the focus in recent years, but the pace of discovery of related genes and their functions in the control of wood properties has been slow. The microarray technique—with its advantages of high throughput capacity, sensitivity, and reliability over other tools developed for investigating genes expression patterns—is capable of rapidly assaying thousands of genes. In this study, a cDNA microarray prepared from two cDNA libraries of developing poplar xylem tissues was used to assay gene expression patterns in immature xylem tissues at different heights from the main stem of Populus deltoides (15 years old), which was confirmed to have distinct wood properties (microfibrillar angle, woody density) by X-ray. Two hundred seventy-four transcripts with differential expression profiles between the chips were screened out, and the individual clones were subjected to 5′ sequencing. Using bioinformatic analysis, we identified candidate genes that may influence poplar wood properties, many of which belong to various regulatory and signal transduction gene families, such as zinc finger protein transcription factor, DNA-binding transcription factor, ethylene response factors, and so on. The results suggest that these genes may regulate enzymes involved in wood formation. Further work will be performed to clone these genes and determine how they influence poplar wood properties.












Similar content being viewed by others
References
Allona I, Quinn M, Shoop E, Swope K, St Cyr SS, Carlis J, Ried J, REtzel E, Campbel MM, Sederoff R, Whetten RW. 1998. Analysis of xylem formation in pine by cDNA sequencing. Proceedings of the National Academy of Sciences USA, 95: 9693-9698
Arioli T, Burn JE, Willianmson RE (2000) Molecular biology of cellulose biosynthesis. In: Jain SM, Minocha SC (eds) Molecular Biology of Woody Plants, vol 1. Kluwer Academic Publishers, Dordrecht, pp 205–226
Chaffey N (2000) Microfibril orientation in wood cells: new angles on an old topic. Trends Plant Sci 5:138–151
Christophe P, Gre´goire L, Alexia S (2001) Wood Formation in Trees. Plant Physiol 127:1513–1523
Esau K (1965) Plant Anatomy. Wiley, New York, p 57
Higuchi T (1997) Biochemistry and Molecular Biology of Wood. Springer-Verlag, New York, pp 131–181
Keegstra K, Raikhel N (2001) Plant glycosyltransferases. Curr Opin Plant Biol 4:219–224
Lacombe E, Van Doorsselaere J, Boerjan W, Boudet AM, Grima-Pettenati J (2000) Characterization of Cis-element required for vascular expression of the cinnamoyl CoA reductase gene and for protein-DNA complex formation. Plant J 23:663–676
Meylan BA (1968) Cause of high longitudinal shrinkage in wood. For Prod J 18(4):75–78
Meylan BA (1972) The influence of microfibril angle on the longitudinal shrinkage-moisture content relationship. Wood Sci Technol 4(4):293–301
Nieminen KM, Kauppinen L, Helariutta Y (2004) A weed for wood? Arabidopsis as a genetic model for xylem development. Plant Physiol 135:653–659
Raemodonck DV, Pesquet E, Cloquet S, Beeckman H, Boerjan W, Goffner D, Jaziri M, Baucher M (2005) Molecular changes associated with the setting up of secondary growth in aspen. J Exp Bot 56(418):2211–2227
Sundberg B, Uggla C, Tuominen H (2000) Cambial growth and auxin gradient. In: Savidge RA, Barnett JR, Napier R (eds) Cell and molecular biology of wood formation. Bios Scientific, Oxford, pp 169–188
Takele D, Robert K. Shepard.1999. Wood properties of red pine. Maine Agricultural and Forest Experiment station Miscellaneous Report 412
Timell TE (1986) Compression Wood in Gymnosperms. Springer-Verlag, Heidelberg, pp 157–167
Zhong R, Burk DH, Morrison WH, Ye ZH (2002) A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 12:3101–3117
Author information
Authors and Affiliations
Corresponding author
Additional information
Project funding
This work was supported by the State Key Basic Research Program of China (No. 2012CB114506).
The online version is available at http://www.springerlink.com
Corresponding editor: Hu Yanbo
Qinjun Huang and Changjun Ding have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Huang, Q., Ding, C., Zhang, W. et al. A cDNA microarray analysis of the molecular control of poplar wood properties. J. For. Res. 28, 71–82 (2017). https://doi.org/10.1007/s11676-016-0298-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-016-0298-y