Abstract
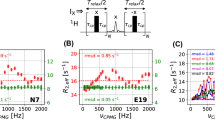
In solution, the correlation time of the overall protein tumbling, τ R , plays a role of a natural dynamics cutoff—internal motions with correlation times on the order of τ R or longer cannot be reliably identified on the basis of spin relaxation data. It has been proposed some time ago that the ‘observation window’ of solution experiments can be expanded by changing the viscosity of solvent to raise the value of τ R . To further explore this concept, we prepared a series of samples of α-spectrin SH3 domain in solvent with increasing concentration of glycerol. In addition to the conventional 15N labeling, the protein was labeled in the Val, Leu methyl positions (13CHD2 on a deuterated background). The collected relaxation data were used in asymmetric fashion: backbone 15N relaxation rates were used to determine τ R across the series of samples, while methyl 13C data were used to probe local dynamics (side-chain motions). In interpreting the results, it has been initially suggested that addition of glycerol leads only to increases in τ R , whereas local motional parameters remain unchanged. Thus the data from multiple samples can be analyzed jointly, with τ R playing the role of experimentally controlled variable. Based on this concept, the extended model-free model was constructed with the intent to capture the effect of ns time-scale rotameric jumps in valine and leucine side chains. Using this model, we made a positive identification of nanosecond dynamics in Val-23 where ns motions were already observed earlier. In several other cases, however, only tentative identification was possible. The lack of definitive results was due to the approximate character of the model—contrary to what has been assumed, addition of glycerol led to a gradual ‘stiffening’ of the protein. This and other observations also shed light on the interaction of the protein with glycerol, which is one of the naturally occurring osmoprotectants. In particular, it has been found that the overall protein tumbling is controlled by the bulk solvent, and not by a thin solvation layer which contains a higher proportion of water.











Similar content being viewed by others
Notes
An additional complication arises from the fact that CspB is thoroughly affected by ms time scale exchange arising from the folding–unfolding equilibrium that exists in this marginally stable protein. Addition of ethylene glycol shifts this equilibrium toward the folded form and thus alters the R ex rates.
In order to extrapolate the results toward the limit of zero glycerol concentration the authors relied on the data from translational diffusion measurements. It should be pointed out that these measurements can be technically demanding; their accuracy is typically much lower than that of the standard τ R determination (Price 1998). In addition, a simple relationship between the translational and rotational diffusion coefficients, in principle, can be violated as a result of the solvent partition (i.e. preferential hydration of the protein surface).
References
Agarwal V, Reif B (2008) Residual methyl protonation in perdeuterated proteins for multi-dimensional correlation experiments in MAS solid-state NMR spectroscopy. J Magn Reson 194:16–24
Agarwal V, Xue Y, Reif B, Skrynnikov NR (2008) Protein side-chain dynamics as observed by solution- and solid-state NMR spectroscopy: a similarity revealed. J Am Chem Soc 130:16611–16621
Akaike H (1973) Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csaki F (eds) Second international symposium on information theory. Akademiai Kiado, Budapest, 267–281
Albertyn J, Hohmann S, Thevelein JM, Prior BA (1994) Gpd1, which encodes gycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol 14:4135–4144
Allison SA, Tran VT (1995) Modeling the electrophoresis of rigid polyions - application to lysozyme. Biophys J 68:2261–2270
Batchelder LS, Sullivan CE, Jelinski LW, Torchia DA (1982) Characterization of leucine side-chain reorientation in collagen fibrils by solid-state 2H NMR. Proc Natl Acad Sci USA 79:386–389
Baynes BM, Trout BL (2003) Proteins in mixed solvents: a molecular-level perspective. J Phys Chem B 107:14058–14067
Ben-Amotz A, Avron M (1973) The role of glycerol in osmotic regulation of the halophilic alga Dunaliella Parva. Plant Physiol 51:875–878
Best RB, Clarke J, Karplus M (2005) What contributions to protein side-chain dynamics are probed by NMR experiments? A molecular dynamics simulation analysis. J Mol Biol 349:185–203
Betting H, Häckel M, Hinz HJ, Stockhausen M (2001) Spectroscopic evidence for the preferential hydration of RNase a in glycerol–water mixtures: dielectric relaxation studies. Phys Chem Chem Phys 3:1688–1692
Bizzarri AR, Cannistraro S (2002) Molecular dynamics of water at the protein-solvent interface. J Phys Chem B 106:6617–6633
Blanco FJ, Ortiz AR, Serrano L (1997) 1H and 15N NMR assignment and solution structure of the SH3 domain of spectrin: comparison of unrefined and refined structure sets with the crystal structure. J Biomol NMR 9:347–357
Burnham KP, Anderson DR (2002) Model selection and multimodel interference: a practical information-theoretic approach. Springer, New York
Butler SL, Falke JJ (1996) Effects of protein stabilizing agents on thermal backbone motions: a disulfide trapping study. Biochemistry 35:10595–10600
Caliskan G, Mechtani D, Roh JH, Kisliuk A, Sokolov AP, Azzam S, Cicerone MT, Lin-Gibson S, Peral I (2004) Protein and solvent dynamics: how strongly are they coupled? J Chem Phys 121:1978–1983
Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H (2002) Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 420:98–102
Charron C, Kadri A, Robert MC, Giege R, Lorber B (2002) Crystallization in the presence of glycerol displaces water molecules in the structure of thaumatin. Acta Crystallogr D Biol Crystallogr 58:2060–2065
Chemical Rubber Company (1948) Handbook of chemistry and physics. Chemical Rubber Publishing, Cleveland
Chen JH, Brooks CL, Wright PE (2004) Model-free analysis of protein dynamics: assessment of accuracy and model selection protocols based on molecular dynamics simulation. J Biomol NMR 29:243–257
Chevelkov V, Faelber K, Diehl A, Heinemann U, Oschkinat H, Reif B (2005) Detection of dynamic water molecules in a microcrystalline sample of the SH3 domain of α-spectrin by MAS solid-state NMR. J Biomol NMR 31:295–310
Chevelkov V, Zhuravleva AV, Xue Y, Reif B, Skrynnikov NR (2007) Combined analysis of 15N relaxation data from solid- and solution-state NMR spectroscopy. J Am Chem Soc 129:12594–12595
Clarkson MW, Lee AL (2004) Long-range dynamic effects of point mutations propagate through side chains in the serine protease inhibitor eglin c. Biochemistry 43:12448–12458
Clore GM, Driscoll PC, Wingfield PT, Gronenborn AM (1990a) Analysis of the backbone dynamics of interleukin 1β using two-dimensional inverse detected heteronuclear 15N–1H NMR spectroscopy. Biochemistry 29:7387–7401
Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM (1990b) Deviations from the simple two-parameter model-free approach to the interpretation of 15N nuclear magnetic relaxation of proteins. J Am Chem Soc 112:4989–4991
Cole R, Loria JP (2003) FAST-modelfree: a program for rapid automated analysis of solution NMR spin-relaxation data. J Biomol NMR 26:203–213
Cornilescu G, Bax A (2000) Measurement of proton, nitrogen, and carbonyl chemical shielding anisotropies in a protein dissolved in a dilute liquid crystalline phase. J Am Chem Soc 122:10143–10154
Courtenay ES, Capp MW, Anderson CF, Record MT (2000) Vapor pressure osmometry studies of osmolyte-protein interactions: Implications for the action of osmoprotectants in vivo and for the interpretation of “osmotic stress” experiments in vitro. Biochemistry 39:4455–4471
Dastidar SG, Mukhopadhyay C (2003) Structure, dynamics, and energetics of water at the surface of a small globular protein: a molecular dynamics simulation. Phys Rev E 68
de la Torre JG, Huertas ML, Carrasco B (2000a) Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J 78:719–730
de la Torre JG, Huertas ML, Carrasco B (2000b) HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J Magn Reson B 147:138–146
Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE (1994) Backbone dynamics of a free and a phosphopeptide-complexed src homology two domain studied by 15N NMR relaxation. Biochemistry 33:5984–6003
Feder ME, Burggren WW (eds) (1992) Environmental physiology of the amphibians. University of Chicago Press, Chicago
Fenimore PW, Frauenfelder H, McMahon BH, Parak FG (2002) Slaving: solvent fluctuations dominate protein dynamics and functions. Proc Natl Acad Sci USA 99:16047–16051
Finerty PJ, Muhandiram R, Forman-Kay JD (2002) Side-chain dynamics of the SAP SH2 domain correlate with a binding hot spot and a region with conformational plasticity. J Mol Biol 322:605–620
Foord RL, Leatherbarrow RJ (1998) Effect of osmolytes on the exchange rates of backbone amide protons in proteins. Biochemistry 37:2969–2978
Frederick KK, Marlow MS, Valentine KG, Wand AJ (2007) Conformational entropy in molecular recognition by proteins. Nature 448:325–329
Garman EF, Doublie S (2003) Cryocooling of macromolecular crystals: optimization methods. Methods Enzymol 368:188–216
Gekko K, Timasheff SN (1981a) Mechanism of protein stabilization by glycerol - preferential hydration in glycerol–water mixtures. Biochemistry 20:4667–4676
Gekko K, Timasheff SN (1981b) Thermodynamic and kinetic examination of protein stabilization by glycerol. Biochemistry 20:4677–4686
Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE (1999) A robust and cost-effective method for the production of Val, Leu, Ile-δ1 methyl-protonated 15N, 13C, 2H-labeled proteins. J Biomol NMR 13:369–374
Hansen DF, Kay LE (2007) Improved magnetization alignment schemes for spin-lock relaxation experiments. J Biomol NMR 37:245–255
Hologne M, Faelber K, Diehl A, Reif B (2005) Characterization of dynamics of perdeuterated proteins by MAS solid-state NMR. J Am Chem Soc 127:11208–11209
Hu H, Clarkson MW, Hermans J, Lee AL (2003) Increased rigidity of eglin c at acidic pH: evidence from NMR spin relaxation and MD simulations. Biochemistry 42:13856–13868
Hu H, Hermans J, Lee AL (2005) Relating side-chain mobility in proteins to rotameric transitions: insights from molecular dynamics simulations and NMR. J Biomol NMR 32:151–162
Hurvich CM, Tsai CL (1989) Regression and time-series model selection in small samples. Biometrika 76:297–307
Igumenova TI, Lee AL, Wand AJ (2005) Backbone and side chain dynamics of mutant calmodulin-peptide complexes. Biochemistry 44:12627–12639
Ishima R, Petkova AP, Louis JM, Torchia DA (2001) Comparison of methyl rotation axis order parameters derived from model-free analyses of 2H and 13C longitudinal and transverse relaxation rates measured in the same protein sample. J Am Chem Soc 123:6164–6171
Johnson E, Chazin WJ, Rance M (2006) Effects of calcium binding on the side-chain methyl dynamics of calbindin D9k: a 2H NMR relaxation study. J Mol Biol 357:1237–1252
Kay LE, Muhandiram DR, Farrow NA, Aubin Y, Forman-Kay JD (1996) Correlation between dynamics and high affinity binding in an SH2 domain interaction. Biochemistry 35:361–368
Kempkes R, Stofko E, Lam K, Snell EH (2008) Glycerol concentrations required for the successful vitrification of cocktail conditions in a high-throughput crystallization screen. Acta Crystallogr D Biol Crystallogr 64:287–301
Knubovets T, Osterhout JJ, Connolly PJ, Klibanov AM (1999) Structure, thermostability, and conformational flexibility of hen egg-white lysozyme dissolved in glycerol. Proc Natl Acad Sci USA 96:1262–1267
Korchuganov DS, Gagnidze IE, Tkach EN, Schulga AA, Kirpichnikov MP, Arseniev AS (2004) Determination of protein rotational correlation time from NMR relaxation data at various solvent viscosities. J Biomol NMR 30:431–442
Korzhnev DM, Skrynnikov NR, Millet O, Torchia DA, Kay LE (2002) An NMR experiment for the accurate measurement of heteronuclear spin-lock relaxation rates. J Am Chem Soc 124:10743–10753
Lee LK, Rance M, Chazin WJ, Palmer AG (1997) Rotational diffusion anisotropy of proteins from simultaneous analysis of N-15 and C-13(alpha) nuclear spin relaxation. J Biomol NMR 9:287–298
Li YC, Montelione GT (1995) Human type-alpha transforming growth-factor undergoes slow conformational exchange between multiple backbone conformations as characterized by 15N relaxation measurements. Biochemistry 34:2408–2423
Lienin SF, Brüschweiler R, Ernst RR (1998) Rotational motion of a solute molecule in a highly viscous liquid studied by 13C NMR: 1, 3-dibromoadamantane in polymeric chlorotrifluoroethene. J Magn Reson 131:184–190
Lipari G, Szabo A (1982a) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc 104:4546–4559
Lipari G, Szabo A (1982b) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc 104:4559–4570
Lovell SC, Word JM, Richardson JS, Richardson DC (2000) The penultimate rotamer library. Proteins Struct Funct Genet 40:389–408
Mandel AM, Akke M, Palmer AG (1995) Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol 246:144–163
Millet O, Muhandiram DR, Skrynnikov NR, Kay LE (2002) Deuterium spin probes of side-chain dynamics in proteins. 1. Measurement of five relaxation rates per deuteron in 13C-labeled and fractionally 2H-enriched proteins in solution. J Am Chem Soc 124:6439–6448
Millet O, Mittermaier A, Baker D, Kay LE (2003) The effects of mutations on motions of side-chains in protein L studied by 2H NMR dynamics and scalar couplings. J Mol Biol 329:551–563
Mittermaier A, Kay LE (2004) The response of internal dynamics to hydrophobic core mutations in the SH3 domain from the fyn tyrosine kinase. Protein Sci 13:1088–1099
Modig K, Liepinsh E, Otting G, Halle B (2004) Dynamics of protein and peptide hydration. J Am Chem Soc 126:102–114
Musacchio A, Noble M, Pauptit R, Wierenga R, Saraste M (1992) Crystal structure of a src-homology 3 (SH3) domain. Nature 359:851–855
Nicholson LK, Kay LE, Baldisseri DM, Arango J, Young PE, Bax A, Torchia DA (1992) Dynamics of methyl groups in proteins as studied by proton-detected 13C NMR spectroscopy: application to the leucine residues of staphylococcal nuclease. Biochemistry 31:5253–5263
Pal SK, Peon J, Zewail AH (2002) Biological water at the protein surface: dynamical solvation probed directly with femtosecond resolution. Proc Natl Acad Sci USA 99:1763–1768
Pegg DE (2007) Principles of cryopreservation. Methods Mol Biol 368:39–57
Priev A, Almagor A, Yedgar S, Gavish B (1996) Glycerol decreases the volume and compressibility of protein interior. Biochemistry 35:2061–2066
Rariy RV, Klibanov AM (1997) Correct protein folding in glycerol. Proc Natl Acad Sci USA 94:13520–13523
Reif B, Xue Y, Agarwal V, Pavlova MS, Hologne M, Diehl A, Ryabov YE, Skrynnikov NR (2006) Protein side-chain dynamics observed by solution- and solid-state NMR: comparative analysis of methyl 2H relaxation data. J Am Chem Soc 128:12354–12355
Roche CJ, Guo F, Friedman JM (2006) Molecular level probing of preferential hydration and its modulation by osmolytes through the use of pyranine complexed to hemoglobin. J Biol Chem 281:38757–38768
Sinibaldi R, Ortore MG, Spinozzi F, Carsughi F, Frielinghaus H, Cinelli S, Onori G, Mariani P (2007) Preferential hydration of lysozyme in water/glycerol mixtures: a small-angle neutron scattering study. J Chem Phys 126
Skrynnikov NR (2007) Asymmetric doublets in MAS NMR: coherent and incoherent mechanisms. Magn Reson Chem 45:S161–S173
Skrynnikov NR, Millet O, Kay LE (2002) Deuterium spin probes of side-chain dynamics in proteins. 2. Spectral density mapping and identification of nanosecond time-scale side-chain motions. J Am Chem Soc 124:6449–6460
Sousa R (1995) Use of glycerol, polyols and other protein structure stabilizing agents in protein crystallization. Acta Crystallogr D Biol Crystallogr 51:271–277
Tarek M, Tobias DJ (2008) The role of protein-solvent hydrogen bond dynamics in the structural relaxation of a protein in glycerol versus water. Eur Biophys J Biophys Lett 37:701–709
Tjandra N, Feller SE, Pastor RW, Bax A (1995) Rotational diffusion anisotropy of human ubiquitin from N-15 NMR relaxation. J Am Chem Soc 117:12562–12566
Tjandra N, Wingfield P, Stahl S, Bax A (1996) Anisotropic rotational diffusion of perdeuterated HIV protease from N-15 NMR relaxation measurements at two magnetic fields. J Biomol NMR 8:273–284
Tollinger M, Skrynnikov NR, Mulder FAA, Forman-Kay JD, Kay LE (2001) Slow dynamics in folded and unfolded states of an SH3 domain. J Am Chem Soc 123:11341–11352
Tugarinov V, Kay LE (2005) Quantitative 13C and 2H NMR relaxation studies of the 723-residue enzyme malate synthase G reveal a dynamic binding interface. Biochemistry 44:15970–15977
Tugarinov V, Kanelis V, Kay LE (2006) Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc 1:749–754
Venable RM, Pastor RW (1988) Frictional models for stochastic simulations of proteins. Biopolymers 27:1001–1014
Woessner DE (1962) Nuclear spin relaxation in ellipsoids undergoing rotational brownian motion. J Chem Phys 37:647–654
Xue Y, Pavlova MS, Ryabov YE, Reif B, Skrynnikov NR (2007) Methyl rotation barriers in proteins from 2H relaxation data. Implications for protein structure. J Am Chem Soc 129:6827–6838
Zeeb M, Jacob MH, Schindler T, Balbach J (2003) 15N relaxation study of the cold shock protein CspB at various solvent viscosities. J Biomol NMR 27:221–234
Acknowledgments
We are grateful to Bernd Reif, Veniamin Chevelkov, and Vipin Agrawal for continuing fruitful collaboration on α-spc SH3. The research has been funded through the NSF grants MCB-044563 and CHE-0723718.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, J., Xue, Y. & Skrynnikov, N.R. Detection of nanosecond time scale side-chain jumps in a protein dissolved in water/glycerol solvent. J Biomol NMR 45, 57–72 (2009). https://doi.org/10.1007/s10858-009-9336-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-009-9336-9