Abstract
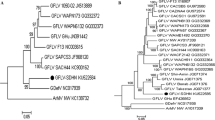
Rice stripe virus (RSV) is one of the most destructive pathogens of rice plants in East Asia. The RSV genome consists of four single-stranded RNA segments. We have determined and compared the complete nucleotide sequences of the RNA1 and RNA2 segments and the deduced amino acid sequence of each ORF of the 13 Korean RSV isolates and established their relationships with reported RSV sequences from China and Japan. Our results showed that the average percent nucleotide divergence based on the full-length genome is higher in RNA2 (2.2%) than in RNA1 (2.0%). The average percent amino acid variation of the RNA-dependent RNA polymerase (RdRp), glycoprotein and NS2 genes encoded by viral complementary (vc) RNA1, viral RNA2 and vcRNA2, showed 2.8, 2.5 and 6.46%, respectively. On the other hand, the average percent nucleotide variation in the intergenic region (IGR) of RNA2 among the 13 Korean-RSV isolates was 3.5%. Phylogenetic analysis of the 13 Korean, 1 Japanese and 5 Chinese isolates based on their complete nucleotide sequences revealed two distinct types of RNA1 and three distinct types of RNA2. Most Chinese isolates grouped with one of the RNA1 types, but they were distributed among the three types when grouped by RNA2. Japanese isolate T was grouped with Korean isolates into one of the RNA1 and RNA2 genotypes. Taken together, our results suggest that the RSV population in Korea consists of mixtures of RNA1–RNA4 genome segments originating from distinctive ancestors, most likely due to either reassortment or recombination events among isolates.


Similar content being viewed by others
References
Falk BW, Tsai JH (1998) Biology and molecular biology of viruses in the genus tenuiviruses. Annu Rev Phytopathol 36:139–163
Felsenstein J (1993) PHYLIP (Phylogeny Interference Package) 3.5. Department of Genetics. University of Washington, Seattle
Garcial-Arenal F, Fraile A (2003) Variation and evolution of plant virus populations. Int Microbiol 6:225–232
Haenni A-L, de Miranda JR, Falk BW, Goldbach R, Mayo MA, Shirako Y, Toriyama S (2005) Tenuivirus. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy: eighth report of the International Committee on taxonomy of viruses. Elsevier, San Diego, pp 717–723
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hamamatsu C, Toriyama S, Toyoda T, Ishihama A (1993) Ambisense coding strategy of the rice stripe virus genome: in vitro translation studies. J Gen Virol 74:1125–1131
Hibino H (1996) Biology and epidemiology of rice viruses. Annu Rev Phytopathol 34:249–274
Jeger MJ, Seal SE, van den Bosch F (2006) Evolutionary epidemiology of plant virus disease. Adv Virus Res 67:163–203
Jonson GM, Choi H-S, Kim J-S, Choi I-R, Kim K-H (2009) Complete genome sequence of the RNAs 3 and 4 segments of Rice stripe virus isolates in Korea and their phylogenetic relationships with Japan and China isolates. Plant Pathol J 25:142–150
Koganezawa H (1977) Purification and properties of rice stripe virus. Trop Agric Res Ser 10:151–154
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russell DW (2001) Molecular cloning, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Toriyama S, Takahashi M, Sano Y, Shimizu T, Ishihama A (1994) Nucleotide sequence of RNA1, the largest genomic segment of Rice stripe virus, the prototype of the tenuivirus. J Gen Virol 75:3569–3579
Toriyama S (2000) Rice stripe virus. CMI/AAB description of plant viruses. No. 375
Wei T-Y, Yang J-G, Liao F-L, Gao F-L, Lu L-M, Zhang X-T, Li F, Wu Z-J, Lin Q-Y, Xie L-H, Lin H-X (2009) Genetic diversity and population structure of rice stripe in China. J Gen Virol 90:1025–1034
Zhang H-M, Yang J, Sun H-R, Xin X, Wang H-D, Chen J-P, Adams MJ (2007) Genomic analysis of Rice stripe virus Zhejiang isolate shows the presence of an OTU-like domain in the RNA1 protein and a novel sequence motif conserved within the intergenic regions of ambisense segments of tenuiviruses. Arch Virol 152:1917–1923
Acknowledgments
This research was supported in part by grants from the Rural Development Administration (No. 20090101-030-144-001), the Korea Science and Engineering Foundation grant (No. R01-2008-062-02003-0) funded by the Ministry of Education, Science and Technology (MEST), and the Agricultural R&D Promotion Center (No. 108103-02-1-SB020). MGJ was supported by a post-doctoral research fellowship from the MEST through the Brain Korea 21 Project.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jonson, M.G., Choi, HS., Kim, JS. et al. Sequence and phylogenetic analysis of the RNA1 and RNA2 segments of Korean Rice stripe virus isolates and comparison with those of China and Japan. Arch Virol 154, 1705–1708 (2009). https://doi.org/10.1007/s00705-009-0493-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-009-0493-7