Abstract
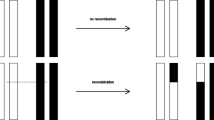
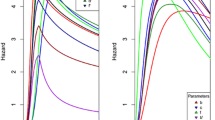
We present a model of gene duplication by means of unequal crossover (UCO) where the probability of any given pairing between homologous sequences scales as a penalty factor p z ≤ 1, with z the number of mismatches due to asymmetric sequence alignment. From this general representation, we derive several limiting case models of UCO, some of which have been treated elsewhere in the literature. One limiting case is random unequal crossover (RUCO), obtained by setting p = 1 (corresponding to equiprobable pairings at each site). Another limiting case scenario (the ‘Krueger-Vogel’ model) proposes an optimal ‘endpoint’ alignment which strongly penalizes both overhang and deviations from endpoint matching positions. For both of these scenarios, we make use of the symmetry properties of the transition operator (together with the more general UCO properties of copy number conservation and equal parent-offspring mean copy number) to derive the stationary distribution of gene copy number generated by UCO. For RUCO, the stationary distribution of genotypes is shown to be a negative binomial, or alternatively, a convolution of geometric distributions on ‘haplotype’ frequencies. A different type of model derived from the general representation only allows recombination without overhang (internal UCO or IntUCO). This process has the special property of converging to a single copy length or a distribution on a pair of copy lengths in the absence of any other evolutionary forces. For UCO systems in general, we also show that selection can readily act on gene copy number in all of the UCO systems we investigate due to the perfect heritability (h 2 = 1) imposed by conservation of copy number. Finally, some preliminary work is presented which suggests that the more general models based on misalignment probabilities seem to also converge to stationary distributions, which are most likely functions of parameter value p.
Similar content being viewed by others
References
Axelrod, D. E., K. A. Baggerly and M. Kimmel (1994). Gene amplification by unequal sister chromatid exchange: probabilistic modeling and analysis of drug resistance data. J. Theor. Biol. 168, 151–159.
Bailey, W. J., J. Kim, G. P. Wagner and F. H. Ruddle (1997). Phylogenetic reconstruction of vertebrate Hox cluster duplications. Mol. Biol. Evol. 14, 843–853.
Barker, W. C., L. C. Ketchum and M. O. Dayhoff (1978). Duplications in protein sequences, in Atlas of Protein Sequences and Structure, Vol. 5, Supplement 3, Silver Springs, MD: National Biomedical Research Foundation, pp. 359–362.
Biebricher, C. K., M. Eigen and T. S. McCaskill (1993). Template directed and template free RNA synthesis. J. Mol. Biol. 231, 175–179.
Biebricher, C. K. and K. Luce (1992). In vitro recombination and terminal elongation of RNA. EMBO J. 11, 5129–5135.
Bowler, M. G. (1982). Lectures on Statistical Mechanics, New York: Pergamon.
Buongiorno-Nardelli, M., F. Amaldi and P. A. Lava-Sanchez (1972). Amplification as a rectification mechanism for redundant rRNA genes. Nat. New Biol. 238, 134.
Charlesworth, B., P. Sniegowski and W. Stephan (1994). The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371, 215–220.
Dayhoff, M. O. (1978). Atlas of Protein Sequences and Structure, Vol. 5, Supplement 3, Silver Springs, MD: National Biomedical Research Foundation.
Du Pasquier, L. (1992). Origin and evolution of the vertebrate immune system. Apmis 100, 383–392.
Durrett, R. and S. Kruglyak (1998). A new stochastic model of microsatellite evolution. J. Appl. Probability 36, 621–631.
Ewens, W. J. (1979). Mathematical Population Genetics, New York: Springer.
Fuchs, E. and C. Byrne (1994). The epidermis: rising to the surface. Curr. Opin. Gen. Dev. 4, 725–736.
Go, M. (1981). Correlation of DNA exonic regions with protein structural units in hemoglobin. Nature 291, 90–92.
Grenier, J. K., T. L. Garber, R. Warren, P. M. Whittington and S. Carroll (1997). Evolution of the entire arthropod Hox gene set predated the origin and radiation of the onychophoran/arthropod clade. Curr. Biol. 7, 547–553.
Haken, H. (1977). Synergetics, New York: Springer.
Houle, D. (1992). Comparing evolvability and variability of quantitative traits. Genetics 130, 195–204.
Huynen, M. and E. van Nimwegen (1998). The frequency distribution of gene family sizes in complete genomes. Mol. Biol. Evol. 15, 583–598.
Krueger, J. and F. Vogel (1975). Population genetics of unequal crossing over. J. Mol. Evol. 4, 201–247.
Kruglyak, S., R. T. Durrett, M. D. Schug and C. F. Aquadro (1998). Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. PNAS 95, 10774–10778.
Lande, R. (1976). Natural selection and random drift in phenotypic evolution. Evolution 30, 314–334.
Lande, R. and S. Arnold (1983). The measurement of selection on correlated characters. Evolution 37, 1210–1226.
Li, W.-H. (1983). Evolution of duplicate genes and pseudogenes, in Evolution of Genes and Proteins, M. Nei and R. K. Koehn (Eds), Sunderland, MA: Sinauer Assoc., pp. 14–37.
Li, W.-H. and D. Grauer (1994). Fundamentals of Molecular Biology, Sunderland, MA: Sinauer Assoc.
Lynch, M. and J. B. Walsh (1996). Genetics and Analysis of Quantitative Traits, Sunderland, MA: Sinauer Assoc.
Maeda, N. and O. Smithies (1986). The evolution of multigene families: human haptoglobin genes. Ann. Rev. Genet. 20, 81–108.
Nagylaki, T. (1984a). The evolution of multigene families under intrachromosomal gene conversion. Genetics 106, 529–548.
Nagylaki, T. (1984b). Evolution of multigene families under intrachromosomal gene conversion. PNAS 81, 3796–3800.
Nowak, M. A., M. C. Boerlist, J. Cooke and J. Maynard Smith (1997). Evolution of genetic redundancy. Nature 388, 167–171.
Ohno, S. (1970). Evolution by Gene Duplication, Berlin: Springer.
Ohta, T. (1980). Functional Variation in Multigene Families, Berlin: Springer-Verlag.
Ohta, T. (1983). On the evolution of multigene families. Theor. Popul. Biol. 23, 216–240.
Ohta, T. (1987). Simulating evolution by gene duplication. Genetics 115, 207–213.
Ohta, T. and G. Dover (1980). Population genetics of multigene families that are dispersed into two or more chromosomes. PNAS 80, 4079–4083.
Pendleton, J. W., B. K. Nagai, M. T. Murtha and F. H. Ruddle (1993). Expansion of the Hox gene family and the evolution of chordates. PNAS 90, 6300–6304.
Perelson, A. S. and G. Bell (1977). Mathematical models for the evolution of multigene families by unequal crossing over. Nature 265, 304–310.
Rabani, Y., Y. Rabinovich and A. Sinclair (1995). A computational view of population genetics, Proceedings of the 27th ACM Symposium on the Theory of Computing, Las Vegas, NV: pp. 83–92.
Rabinovich, Y., A. Sinclair and A. Wigderson (1992). Quadratic dynamical systems, Proceedings of the 33rd IEEE Symposium on the Foundations of Computer Science, pp. 304–313.
Roughgarden, J. (1979). Theory of Population Genetics and Evolutionary Ecology: An Introduction, New York: Prentice Hall.
Schluter, S. F., E. Schroeder, E. Wang and J. J. Marschalonis (1994). Recognition molecules and immunoglobin domains in invertebrates. Ann. N. Y. Acad. Sci. 712, 74–81.
Shpak, M. and G. P. Wagner (2000). Asymmetry of configuration spaces induced by unequal crossover. Artif. Life 6, 25–43.
Smith, G. P. (1973). Unequal crossover and the evolution of multigene families. Cold Springs Harb. Symp. Quant. Biol. 38, 507–513.
Smith, K. A., P. A. Gorman, M. B. Stark, R. P. Groves and G. P. Stark (1990). Distinctive chromosome structures are formed very early in the amplification of CAD genes in Syrian hamster cells. Cell 63, 1219–1227.
Smith, K. A., M. B. Stark, P. A. Gorman and G. R. Stark (1992). Fusions near telomeres occur very early in the amplification of CAD genes in Syrian hamster cells. PNAS USA 89, 5427–5431.
Smithies, O. (1964). Chromosomal rearrangements and protein structure. Cold Springs Harb. Symp. Quant. Biol. 29, 309–323.
Stadler, B., P. F. Stadler, M. Shpak and G. P. Wagner (2002). Recombination spaces, metrics, and pretopologies. Z. Phys. Chemie 216, 217–234.
Takahata, N. (1981). A mathematical study on the distribution of the number of repeated genes per chromosome. Genet. Res. 38, 97–102.
Walsh, J. B. (1987). Persistence of tandem arrays: implications for satellite and simplesequence DNA’s. Genetics 115, 553–567.
Zimmer, E. A., S. L. Martin, S. M. Beverley, Y. W. Kan and A. C. Wilson (1986). Rapid duplication and loss of genes coding for the α chains of hemoglobin. PNAS 77, 2158–2162.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article is available at http://dx.doi.org/10.1006/bulm.2002.0312.
Rights and permissions
About this article
Cite this article
Shpak, M., Atteson, K. A survey of unequal crossover systems and their mathematical properties. Bull. Math. Biol. 64, 703–746 (2002). https://doi.org/10.1006/bulm.2001.0299
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1006/bulm.2001.0299