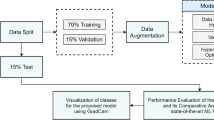
Abstract
In this study, a Convolutional Neural Network (CNN) model for tea leaf disease classification is introduced, which utilizes an encoder–decoder structure with skip connections. The proposed model is designed to reduce computational complexity and generate high-resolution images from lower-resolution representations obtained using the encoder. The skip connections in the model enable the transfer of information between the encoder and decoder, thereby allowing for the reconstruction of the original dimension image from the encoded representation. The performance of the proposed CNN model is compared against several popular computer vision models, including GoogleNet, ResNet50, and VGG16. An ensemble model is also developed to further improve classification performance. In addition, two hybrid CNN models are created, one by combining the computer vision models without using them as feature extractors, and the other by using their feature extraction capabilities with various machine learning algorithms for image classification. Experimental results indicate that the proposed CNN model with skip connections outperforms the individual models with accuracy, precision, recall, and F1-score 87.36, 88.12, 87.36, 87.30, respectively. Based on our experimental results, our proposed CNN model with skip connections did not outperform the ensemble and hybrid models in terms of accuracy. However, the difference in accuracy was marginal, and our proposed model offered lower computational complexity, making it a more efficient option. Therefore, while our proposed model may not be the best performer in terms of accuracy, it can still provide a viable alternative for tea leaf disease classification, especially when computational efficiency is a critical factor.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
International tea market: market situation, prospects, and emerging issues, food and agriculture organization of the United Nations (FAO), Published: May 2022
Chen J et al (2020) Automatic recognition of tea diseases based on deep learning. Adv Manage Global Change (IntechOpen) 2020 chapter 8. https://doi.org/10.5772/intechopen.91953
Bao W, Fan T, Hu G et al (2022) Detection and identification of tea leaf diseases based on AX-RetinaNet. Sci Rep 12:2183. https://doi.org/10.1038/s41598-022-06181-z
Xie S et al (2023) Online identification method of tea diseases in complex natural environments. IEEE Open J Comput Soc 4. https://doi.org/10.1109/OJCS.2023.3247505
Jayapal SK et al (2023) Enhanced disease identification model for tea plant using deep learning. Intell Autom Soft Comput. https://doi.org/10.32604/iasc.2023.026564
Pandian JA et al (2023) Grey blight disease detection on tea leaves using improved deep convolutional neural network. Comput Intell Neurosci 2023. https://doi.org/10.1155/2023/7876302
Javidan SM et al (2023) Diagnosis of grape leaf diseases using automatic K-means clustering and machine learning. Smart Agric Technol 3:100081. https://doi.org/10.1016/j.atech.2022.100081
Gayathri S et al (2020) Image analysis and detection of tea leaf disease using deep learning. In: 2020 international conference on electronics and sustainable communication systems (ICESC). https://doi.org/10.1109/ICESC48915.2020.9155850
Hu et al (2022) Using a multi-convolutional neural network to automatically identify small-sample tea leaf diseases. Sustain Comput Inf Syst 35:100696. https://doi.org/10.1016/j.suscom.2022.100696
Latha RS et al (2021) Automatic detection of tea leaf diseases using deep convolution neural network. In: 2021 international conference on computer communication and informatics (ICCCI). https://doi.org/10.1109/ICCCI50826.2021.9402225
Hu G et al (2019) A low shot learning method for tea leaf’s disease identification. Comput Electron Agri 163:104852. https://doi.org/10.1016/j.compag.2019.104852
Xiaoxiao SUN (2018) Image recognition of tea leaf diseases based on convolutional neural network. In: 2018 international conference on security, pattern analysis, and cybernetics (SPAC). https://doi.org/10.1109/SPAC46244.2018.8965555
Ramadan A (2020) Transfer learning and fine-tuning for deep learning-based tea diseases detection on small datasets. In: Ade ramadan, transfer learning and fine-tuning for deep learning-based tea diseases detection on small datasets. https://doi.org/10.1109/ICRAMET51080.2020.9298575,10.1109/ICRAMET51080.2020.9298575
Hu G et al (2021) Detection and severity analysis of tea leaf blight based on deep learning. Comput Electr Eng 90:107023. https://doi.org/10.1016/j.compeleceng.2021.107023
Chen J et al (2019) Visual tea leaf disease recognition using a convolutional neural network model. Symmetry 11(3):343. https://doi.org/10.3390/sym11030343
Kaur P et al (2022) An approach for characterization of infected area in tomato leaf disease based on deep learning and object detection technique. Eng Appl Artif Intell 115:105210. https://doi.org/10.1016/j.engappai.2022.105210
Kimutai G, Förster A (2022) Tea sickness dataset. Mendeley Data V2. https://doi.org/10.17632/j32xdt2ff5.2
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Shinde, S., Lahade, S. (2023). Tea Leaf Disease Classification Using an Encoder-Decoder Convolutional Neural Network with Skip Connections. In: Shakya, S., Tavares, J.M.R.S., Fernández-Caballero, A., Papakostas, G. (eds) Fourth International Conference on Image Processing and Capsule Networks. ICIPCN 2023. Lecture Notes in Networks and Systems, vol 798. Springer, Singapore. https://doi.org/10.1007/978-981-99-7093-3_24
Download citation
DOI: https://doi.org/10.1007/978-981-99-7093-3_24
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-7092-6
Online ISBN: 978-981-99-7093-3
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)