Abstract
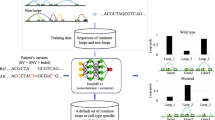
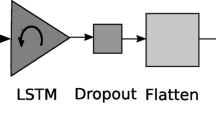
YY1-mediated chromatin loops play substantial roles in basic biological processes like gene regulation, cell differentiation, and DNA replication. YY1-mediated chromatin loop prediction is important to understand diverse types of biological processes which may lead to the development of new therapeutics for neurological disorders and cancers. Existing deep learning predictors are capable to predict YY1-mediated chromatin loops in two different cell lines however, they showed limited performance for the prediction of YY1-mediated loops in the same cell lines and suffer significant performance deterioration in cross cell line setting. To provide computational predictors capable of performing large-scale analyses of YY1-mediated loop prediction across multiple cell lines, this paper presents two novel deep learning predictors. The two proposed predictors make use of Word2vec, one hot encoding for sequence representation and long short-term memory, and a convolution neural network along with a gradient flow strategy similar to DenseNet architectures. Both of the predictors are evaluated on two different benchmark datasets of two cell lines HCT116 and K562. Overall the proposed predictors outperform existing DEEPYY1 predictor with an average maximum margin of 4.65%, 7.45% in terms of AUROC, and accuracy, across both of the datases over the independent test sets and 5.1%, 3.2% over 5-fold validation. In terms of cross-cell evaluation, the proposed predictors boast maximum performance enhancements of up to 9.5% and 27.1% in terms of AUROC over HCT116 and K562 datasets.
This work was supported by Sartorius Artificial Intelligence Lab.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Hepler, J.R., Gilman, A.G.: G proteins. Trends Biochem. Sci. 17(10), 383–387 (1992)
Bonev, B., Cavalli, G.: Organization and function of the 3d genome. Nat. Rev. Genet. 17(11), 661–678 (2016)
Dixon, J.R.: Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485(7398), 376–380 (2012)
He, Y., Casaccia-Bonnefil, P.: The yin and yang of yy1 in the nervous system. J. Neurochem. 106(4), 1493–1502 (2008)
Carminho-Rodrigues, M.T., et al.: Complex movement disorder in a patient with heterozygous yy1 mutation (gabriele-de vries syndrome). Am. J. Med. Genet. Part A 182(9), 2129–2132 (2020)
Verheul, T.C.J., van Hijfte, L., Perenthaler, E., Barakat, T.S.: The why of yy1: mechanisms of transcriptional regulation by yin yang 1. Frontiers in cell and developmental biology, p. 1034 (2020)
Wang, R., Wang, Y., Zhang, X., Zhang, Y., Xiaoyong, D., Fang, Y., Li, G.: Hierarchical cooperation of transcription factors from integration analysis of dna sequences, chip-seq and chia-pet data. BMC Genomics 20(3), 1–13 (2019)
Leina, L., Sun, K., Xiaona Chen, Yu., Zhao, L.W., Zhou, L., Sun, H., Wang, H.: Genome-wide survey by chip-seq reveals yy1 regulation of lincrnas in skeletal myogenesis. EMBO J. 32(19), 2575–2588 (2013)
Kan, S.L., Saksouk, N., Déjardin, J.: Proteome characterization of a chromatin locus using the proteomics of isolated chromatin segments approach. In Proteomics, pp. 19–33. Springer (2017)
Lv, H., Dao, F.-Y., Zulfiqar, H., Su, W., Ding, H., Liu, L., Lin, H.: A sequence-based deep learning approach to predict ctcf-mediated chromatin loop. Briefings Bioinform. 22(5), bbab031 (2021)
Cao, F., et al.: Chromatin interaction neural network (chinn): a machine learning-based method for predicting chromatin interactions from dna sequences. Genome Biol. 22(1), 1–25 (2021)
Dao, F.-Y., Lv, H., Zhang, D., Zhang, Z.-M., Liu, L., Lin, H.: Deepyy1: a deep learning approach to identify yy1-mediated chromatin loops. Briefings in bioinformatics, 22(4):bbaa356, 2021
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Albawi, S., Mohammed, T.A., Al-Zawi, S.: Understanding of a convolutional neural network. In: 2017 International Conference on Engineering and Technology (ICET), pp. 1–6. IEEE (2017)
Kenneth Ward Church: Word2vec. Nat. Lang. Eng. 23(1), 155–162 (2017)
Zhang, R., Wang, Y., Yang, Y., Zhang, Y., Ma, J.: Predicting ctcf-mediated chromatin loops using ctcf-mp. Bioinformatics 34(13), i133–i141 (2018)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Weintraub, A.S., et al.: Yy1 is a structural regulator of enhancer-promoter loops. Cell 171(7), 1573–1588 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Abbasi, A.F., Asim, M.N., Trygg, J., Dengel, A., Ahmed, S. (2023). Deep Learning Architectures for the Prediction of YY1-Mediated Chromatin Loops. In: Guo, X., Mangul, S., Patterson, M., Zelikovsky, A. (eds) Bioinformatics Research and Applications. ISBRA 2023. Lecture Notes in Computer Science(), vol 14248. Springer, Singapore. https://doi.org/10.1007/978-981-99-7074-2_6
Download citation
DOI: https://doi.org/10.1007/978-981-99-7074-2_6
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-7073-5
Online ISBN: 978-981-99-7074-2
eBook Packages: Computer ScienceComputer Science (R0)