Abstract
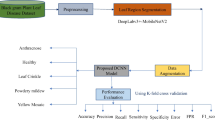
Identifying plant diseases is typically difficult without the assistance of professionals. The disease disrupts the natural growth of the plant. Sometimes, the disease partially infects the plant; sometimes, it infects the whole plant. Several researchers around the world have proposed neural network-based solutions for classifying plant diseases. This research paper presents a method for identifying jute leaf disease (JuLeDI) by utilizing a four-layer-based convolutional neural network (CNN). The CNN model consists of four convolutional layers and four max-pooling layers, followed by two fully connected layers. The CNN-based model was trained using a dataset of 4740 jute leaf images collected from various jute fields in Manikganj, Bangladesh. The dataset included 1600 leaf images of yellow mosaic disease, 1540 leaf images of powdery mildew disease, and 1600 images of healthy jute leaves. The images were captured using a mobile camera. Our proposed convolutional neural network (CNN) model was able to achieve an average accuracy of 96% in classifying jute leaves into three classes: two disease classes (yellow mosaic and powdery mildew) and a healthy class. Compared to state-of-the-art methods such as GPDCNN and SVM, the proposed CNN model achieves the highest accuracy in identifying jute leaf diseases.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Choudhary SB, Sharma HK, Karmakar PG, Kumar AA, Saha AR, Hazra P, Mahapatra BS (2013) Nutritional profile of cultivated and wild jute (‘corchorus’) species. Aust J Crop Sci 7(13):1973–1982
The-Business-Standard. Jute exports rise record 31% in fy21. https://www.tbsnews.net/economy/jute-exports-rise-record-31-fy21-291742
Export Promotion Bureau, B. Export statistics reports. http://www.epb.gov.bd
De RK. Jute diseases: diagnosis and management. https://crijaf.icar.gov.in/pdf/tb_04_2019.pdf
Biswas S (2021) Diseases and pests of fibre crops: ıdentification, treatment and management. Taylor & Francis Group
Sastry KS (2013) Introduction to plant virus and viroid diseases in the tropics. In: Plant virus and viroid diseases in the tropics. Springer, pp 1–10
Ghosh R, Palit P, Paul S, Ghosh SK, Roy A (2012) Detection of corchorus golden mosaic virus associated with yellow mosaic disease of jute (corchorus capsularis). Ind J Virol 23(1):70–74
Das S, Khokon MAR, Haque MM, Ashrafuzzaman M (2001) Jute leaf mosaic and its effects on jute production. Pak J Biol Sci (Pakistan)
Wallelign S, Polceanu M, Buche C (2018) Soybean plant disease identification using convolutional neural network. In: The thirty-first international flairs conference
Wu Q, Chen Y, Meng J (2020) Dcgan-based data augmentation for tomato leaf disease identification. IEEE Access 8:98716–98728
Zhang S, Zhang S, Zhang C, Wang X, Shi Y (2019) Cucumber leaf disease identification with global pooling dilated convolutional neural network. Comput Electron Agric 162:422–430
Reza ZN, Nuzhat F, Mahsa NA, Ali MH (2016) Detecting jute plant disease using image processing and machine learning. In: 2016 3rd ınternational conference on electrical engineering and ınformation communication technology (ICEEICT), pp 1–6
Hasan MZ, Ahamed MS, Rakshit A, Hasan KMZ (2019) Recognition of jute diseases by leaf image classification using convolutional neural network. In: 2019 10th International conference on computing, communication and networking technologies (ICCCNT), pp 1–5
Ashok S, Kishore G, Rajesh V, Suchitra S, Sophia SG, Pavithra B (2020) Tomato leaf disease detection using deep learning techniques. In: 2020 5th International conference on communication and electronics systems (ICCES). IEEE, , pp 979–983
Trivedi J, Shamnani Y, Gajjar R (2020) Plant leaf disease detection using machine learning. In: International conference on emerging technology trends in electronics communication and networking. Springer, pp 267–276
Hughes D, Salathe M et al (2015) An open access repository of images on plant health to enable ´the development of mobile disease diagnostics. arXiv:1511.08060
Surya R, Gautama E (2020) Cassava leaf disease detection using convolutional neural networks. In: 2020 6th International conference on science in ınformation technology (ICSITech), pp 97–102
Taylor D (2014) Work the shell: framing images with imagemagick. Linux J 2014(238):7
Acknowledgements
Several individuals, farmers, and villagers helped us during the data collection process. Data collection was difficult without their cooperation. We sincerely thank them.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendix
Appendix
Leaf diseases of Jute
Jute is an important fiber crop that is grown for the production of burlap, hessian, and twine. However, like any plant, jute is susceptible to a variety of diseases that can significantly reduce crop yield and quality. Here are some common jute diseases and their remedies:
Yellow mosaic is a viral disease that affects jute plants, as well as other plants in the family Tiliaceae, such as mango and linden. It is caused by the jute yellow mosaic virus (JYMV), which is transmitted by aphids. The symptoms of yellow mosaic of jute include yellowing and mottling of the leaves, stunted growth, and reduced yield. The leaves may also develop necrotic lesions and become curled or distorted. There is no cure for yellow mosaic of jute once a plant is infected. The best way to prevent the disease is to control the aphid vectors that transmit the virus. This can be done through the use of chemical insecticides or by using natural predators such as ladybugs, lacewings, and parasitic wasps. In addition, avoiding planting jute in areas where the disease has been previously reported and practicing good sanitation by removing and destroying infected plants can help to prevent the spread of the disease.
Powdery mildew is typically known as a fungal disease that affects the leaves and stems of jute plants. It is characterized by a white or gray powdery growth on the plant’s surface. The fungus that causes powdery mildew is called Oidium juli. Powdery mildew is a common disease of jute, particularly in humid, warm conditions. It can reduce the photosynthetic capacity of the plant and lead to reduced growth and yield. In severe cases, it can cause the death of the plant. To prevent and control powdery mildew of jute, it is important to follow good cultural practices such as proper watering, fertilization, and crop rotation. In addition, it may be necessary to use chemical fungicides to control the disease. Fungicides containing the active ingredients propiconazole or mancozeb are effective at controlling powdery mildew of jute.
Computation of Performance Evaluation Metrics
To evaluate the performance of our CNN model for jute leaf disease identification, we used several assessment metrics such as precision, accuracy, F1-score, and recall. By calculating these metrics for each class, we can better understand how well our model performs in identifying each class. The mathematical expressions describing these metrics are presented below:
Accuracy: Accuracy measures the overall accuracy of the model, taking into account both true positive and true negative predictions. The formula for the accuracy of three class classifications is:
where \({\text{Total}}_{{{\text{Samples}}}}\) represents the total number of sample images. The true positive of Healthy Leaf, Yellow Mosaic, and Powdery Mildew classes are denoted by \({\text{TP}}_{{{\text{HL}}}}\), \({\text{TP}}_{{{\text{YM}}}}\), and \({\text{TP}}_{{{\text{PM}}}} ,\) respectively.
Precision: The precision metric is used to measure the proportion of true positive predictions made by the model compared to the total number of positive predictions. For a three-class classification problem, precision is calculated for each class separately using the following formulas:
where FPHL, FPPM, and FPYM denote the false positive of Healthy Leaf, Powdery Mildew, and Yellow Mosaic, respectively.
Recall: The recall metric evaluates the percentage of true positive predictions among all positive observations in the dataset. It can be calculated using the following formula:
where the value of, i = {Healthy Leaf, Yellow Mosaic, Powdery Mildew}.
The FNi − denotes the false negative of ith class.
F1-Score: The F1-score, which combines precision and recall into a single metric, is often used as a summary statistic for evaluating a model. It is calculated using the following formula:
where the value of, Index = {Healthy Leaf, Yellow Mosaic, Powdery Mildew}. For instance, PrecisionHealthy Leaf—represents the precision value of healthy leaf class.
In the context of multi-class classification with the classes “Healthy leaf,” “Powdery Mildew,” and “Yellow Mosaic,” the following terms have the following meanings:
-
1.
True positive (TP): A sample that is correctly classified as positive for a particular class. For example, if a leaf sample is infected with powdery mildew and the model correctly predicts that it belongs to the “Powdery Mildew” class, then this is a true positive for the “Powdery Mildew” class.
-
2.
True negative (TN): A sample that is correctly classified as negative for a particular class. For example, if a healthy leaf sample is classified as “Healthy leaf” by the model, then this is a true negative for the “Powdery Mildew” and “Yellow Mosaic” classes.
-
3.
False positive (FP): A sample that is incorrectly classified as positive for a particular class. For example, if a healthy leaf sample is misclassified as “Powdery Mildew” by the model, then this is a false positive for the “Powdery Mildew” class.
-
4.
False negative (FN): A sample that is incorrectly classified as negative for a particular class. For example, if a leaf sample infected with powdery mildew is misclassified as “Healthy leaf” by the model, then this is a false negative for the “Powdery Mildew” class.
These metrics are important for evaluating the performance of a multi-class classification model, as they measure the model’s ability to accurately identify samples from each individual class.
Healthy Leaf Class From the confusion matrix, it was found that out of the total 128 samples, 123 samples were correctly classified as belonging to the healthy class.
-
\({\text{Accuracy }} = \left( {123/ 128} \right)\,* \,100\% = 96.00\%\)
-
\({\text{Precision}} = 123/ \left( {123 + 2 + 3} \right) = 0.948\)
-
\({\text{Recall }} = 123/ \left( {123 + 0 + 5} \right) = 0.961\)
-
\({\text{F1-score}} = 2\,*\, \left( {0.948 * 0.961} \right)/ \left( {0.948 + 0.961} \right) = 0.954\)
Eqs. 6, 7, 10, and 11 are used for calculating precision, accuracy, recall, and F1-score, respectively.
Powdery Mildew Class According to the confusion matrix, in the powdery mildew class, the total number of samples is 124; among them 119 samples are correctly identified as powdery mildew.
-
\({\text{Accuracy }} = \left( {119/ 124} \right) * 100\% = 96.96\% = 96.00\%\)
-
\({\text{Precision}} = 119/ \left( {1 + 119 + 4} \right) = 0.956\)
-
\({\text{Recall}} = 119/ \left( {2 + 119 + 3} \right) = 0.952\)
-
\({\text{F1 - score }} = 2 * \left( {0.956 * 0.952} \right) /\left( {0.956 + 0.952} \right) = 0.954\)
Eqs. 6, 8, 10, and 11 are used for calculating precision, accuracy, recall, and F1-score, respectively.
Yellow Mosaic Class According to the confusion matrix, in the yellow mosaic class, the total number of samples is 128; among them 123 samples are correctly identified as powdery mildew.
-
\({\text{Accuracy}} = \left( {123/ 128} \right)\,*\, 100\% = 96.00\%\)
-
\({\text{Precision}} = 124/ \left( {1 + 3 + 124} \right) = 0.968\)
-
\({\text{Recall}} = 124/ \left( {3 + 1 + 124} \right) = 0.971\)
-
\({\text{F1-score}} = 2\,*\,\left( {0.968 * 0.971} \right)/ \left( {0.968 + 0.971} \right) = 0.969\)
Eqs. 6, 9, 10, and 11 are used for calculating precision, accuracy, recall, and F1-score, respectively.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Uddin, M.S., Munsi, M.Y. (2023). JuLeDI: Jute Leaf Disease Identification Using Convolutional Neural Network. In: Ranganathan, G., Papakostas, G.A., Rocha, Á. (eds) Inventive Communication and Computational Technologies. ICICCT 2023. Lecture Notes in Networks and Systems, vol 757. Springer, Singapore. https://doi.org/10.1007/978-981-99-5166-6_45
Download citation
DOI: https://doi.org/10.1007/978-981-99-5166-6_45
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-5165-9
Online ISBN: 978-981-99-5166-6
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)