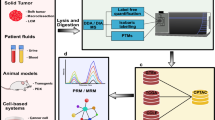
Abstract
Biomarkers factor into the diagnosis and treatment of almost every patient with cancer. The innovation in proteomics follows improvement of mass spectrometry techniques and data processing strategy. Recently, proteomics and typical biological studies have been the answer for clinical applications. The clinical proteomics techniques are now actively adapted to protein identification in large patient cohort, biomarker development for more sensitive and specific screening based on quantitative data. And, it is important for clinical, translational researchers to be acutely aware of the issues surrounding appropriate biomarker development, in order to facilitate entry of clinically useful biomarkers into the clinic. Here, we discuss in detail include the case research for clinical proteomics. Furthermore, we give an overview on the current developments and novel findings in proteomics-based cancer biomarker research.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24(8):971–83.
Gromov P, Moreira JM, Gromova I, Celis JE. Proteomic strategies in bladder cancer: from tissue to fluid and back. Proteomics Clin Appl. 2008;2(7–8):974–88.
Reymond MA, Schlegel W. Proteomics in cancer. Adv Clin Chem. 2007;44:103–42.
Cowan ML, Vera J. Proteomics: advances in biomarker discovery. Expert Rev Proteomics. 2008;5(1):21–3.
Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–67.
Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6(23):6326–53.
Sallam RM. Proteomics in cancer biomarkers discovery: challenges and applications. Dis Markers. 2015;2015:321370.
Chang JW, Kang UB, Kim DH, Yi JK, Lee JW, Noh DY, et al. Identification of circulating endorepellin LG3 fragment: potential use as a serological biomarker for breast cancer. Proteomics Clin Appl. 2008;2(1):23–32.
Francis FL, Jankova L, Dent OF, Molloy MP, Kwun SY, Clarke C, et al. Identification of distinctive protein expression patterns in colorectal adenoma. Proteomics Clin Appl. 2010;4(1):60–70.
Hamelin C, Cornut E, Poirier F, Pons S, Beaulieu C, Charrier JP, et al. Identification and verification of heat shock protein 60 as a potential serum marker for colorectal cancer. FEBS J. 2011;278(24):4845–59.
Magdeldin S, Enany S, Yoshida Y, Xu B, Zhang Y, Zureena Z, et al. Basics and recent advances of two-dimensional- polyacrylamide gel electrophoresis. Clin Proteomics. 2014;11(1):16.
Kim HJ, Kang UB, Lee H, Jung JH, Lee ST, Yu MH, et al. Profiling of differentially expressed proteins in stage IV colorectal cancers with good and poor outcomes. J Proteome. 2012;75(10):2983–97.
Zhou W, Liotta LA, Petricoin EF. The spectra count label-free quantitation in cancer proteomics. Cancer Genomics Proteomics. 2012;9(3):135–42.
Zhu W, Smith JW, Huang CM. Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol. 2010;2010:840518.
Beretov J, Wasinger VC, Millar EK, Schwartz P, Graham PH, Li Y. Proteomic analysis of urine to identify breast cancer biomarker candidates using a label-free LC-MS/MS approach. PLoS One. 2015;10(11):e0141876.
Chen L, Zhao W, He J, Li L, Meng D, Cai D, et al. Label-free quantitative proteomic screening of candidate plasma biomarkers for the prognosis of breast cancer with different lymph node statuses. Proteomics Clin Appl. 2018;12(3):e1700117.
Rudnick PA, Wang X, Yan X, Sedransk N, Stein SE. Improved normalization of systematic biases affecting ion current measurements in label-free proteomics data. Mol Cell Proteomics. 2014;13(5):1341–51.
Nakamura T, Oda Y. Mass spectrometry-based quantitative proteomics. Biotechnol Genet Eng Rev. 2007;24:147–63.
Ciccimaro E, Blair IA. Stable isotope dilution LC-MS for quantitative biomarker analysis. Bioanalysis. 2010;2(2):311–41.
Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–86.
Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21(3):315–8.
Stastna M, van Eyk JE. Investigating the secretome: lessons about the cells that comprise the heart. Circ Cardiovasc Genet. 2012;5(1):o8–o18.
Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods. 2010;7(5):383–5.
Boersema PJ, Geiger T, Wisniewski JR, Mann M. Quantification of the N-glycosylated secretome by super-SILAC during breast cancer progression and in human blood samples. Mol Cell Proteomics. 2013;12(1):158–71.
Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17(10):994–9.
Kang UB, Alexander WM, Marto JA. Interrogating the hidden phosphoproteome. Proteomics. 2017;17(6)
Schneider LV, Hall MP. Stable isotope methods for high-precision proteomics. Drug Discov Today. 2005;10(5):353–63.
Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, et al. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5(11):2167–74.
Yocum AK, Yu K, Oe T, Blair IA. Effect of immunoaffinity depletion of human serum during proteomic investigations. J Proteome Res. 2005;4(5):1722–31.
Brand J, Haslberger T, Zolg W, Pestlin G, Palme S. Depletion efficiency and recovery of trace markers from a multiparameter immunodepletion column. Proteomics. 2006;6(11):3236–42.
Kang UB, Ahn Y, Lee JW, Kim YH, Kim J, Yu MH, et al. Differential profiling of breast cancer plasma proteome by isotope-coded affinity tagging method reveals biotinidase as a breast cancer biomarker. BMC Cancer. 2010;10:114.
DeSouza LV, Taylor AM, Li W, Minkoff MS, Romaschin AD, Colgan TJ, et al. Multiple reaction monitoring of mTRAQ-labeled peptides enables absolute quantification of endogenous levels of a potential cancer marker in cancerous and normal endometrial tissues. J Proteome Res. 2008;7(8):3525–34.
Kang UB, Yeom J, Kim H, Lee C. Quantitative analysis of mTRAQ-labeled proteome using full MS scans. J Proteome Res. 2010;9(7):3750–8.
Kang UB, Yeom J, Kim HJ, Kim H, Lee C. Expression profiling of more than 3500 proteins of MSS-type colorectal cancer by stable isotope labeling and mass spectrometry. J Proteome. 2012;75(10):3050–62.
Yoon JY, Lim KY, Lee S, Park K, Paek E, Kang UB, et al. Improved quantitative analysis of mass spectrometry using quadratic equations. J Proteome Res. 2010;9(5):2775–85.
Suh EJ, Kabir MH, Kang UB, Lee JW, Yu J, Noh DY, et al. Comparative profiling of plasma proteome from breast cancer patients reveals thrombospondin-1 and BRWD3 as serological biomarkers. Exp Mol Med. 2012;44(1):36–44.
Choe L, D'Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, et al. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics. 2007;7(20):3651–60.
Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc. 2018;13(7):1632–61.
Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8(8):645–54.
Labidi-Galy SI, Clauss A, Ng V, Duraisamy S, Elias KM, Piao HY, et al. Elafin drives poor outcome in high-grade serous ovarian cancers and basal-like breast tumors. Oncogene. 2015;34(3):373–83.
Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6(12):2212–29.
Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, Park BK. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(13):1229–39.
Lee HB, Kang UB, Moon HG, Lee J, Lee KM, Yi M, et al. Development and validation of a novel plasma protein signature for breast cancer diagnosis by using multiple reaction monitoring-based mass spectrometry. Anticancer Res. 2015;35(11):6271–9.
Kim DH, Bae J, Lee JW, Kim SY, Kim YH, Bae JY, et al. Proteomic analysis of breast cancer tissue reveals upregulation of actin-remodeling proteins and its relevance to cancer invasiveness. Proteomics Clin Appl. 2009;3(1):30–40.
Kim Y, Kang UB, Kim S, Lee HB, Moon HG, Han W, et al. A validation study of a multiple reaction monitoring-based proteomic assay to diagnose breast cancer. J Breast Cancer. 2019;22(4):579–86.
Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin Chem. 2010;56(2):177–85.
Petushkova NA, Zgoda VG, Pyatnitskiy MA, Larina OV, Teryaeva NB, Potapov AA, et al. Post-translational modifications of FDA-approved plasma biomarkers in glioblastoma samples. PLoS One. 2017;12(5):e0177427.
Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40(Database issue):D261–70.
Ryslava H, Doubnerova V, Kavan D, Vanek O. Effect of posttranslational modifications on enzyme function and assembly. J Proteome. 2013;92:80–109.
Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452(7187):553–63.
Sawyers CL. The cancer biomarker problem. Nature. 2008;452(7187):548–52.
Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–9.
Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7(6):391–403.
Spickett CM, Pitt AR, Morrice N, Kolch W. Proteomic analysis of phosphorylation, oxidation and nitrosylation in signal transduction. Biochim Biophys Acta. 2006;1764(12):1823–41.
Pagel O, Loroch S, Sickmann A, Zahedi RP. Current strategies and findings in clinically relevant post-translational modification-specific proteomics. Expert Rev Proteomics. 2015;12(3):235–53.
Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20(3):301–5.
Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem. 2004;76(14):3935–43.
Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol Cell Proteomics. 2008;7(4):661–71.
Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, et al. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8(2):215–25.
Zhan X, Desiderio DM. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Anal Biochem. 2006;354(2):279–89.
Udeshi ND, Mertins P, Svinkina T, Carr SA. Large-scale identification of ubiquitination sites by mass spectrometry. Nat Protoc. 2013;8(10):1950–60.
Pandey A, Podtelejnikov AV, Blagoev B, Bustelo XR, Mann M, Lodish HF. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc Natl Acad Sci U S A. 2000;97(1):179–84.
Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–6.
Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21(6):667–72.
Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin Chem. 2006;52(10):1897–905.
Ahn Y, Kang UB, Kim J, Lee C. Mining of serum glycoproteins by an indirect approach using cell line secretome. Mol Cells. 2010;29(2):123–30.
Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J Proteome Res. 2005;4(3):998–1005.
Kim S, Na S, Sim JW, Park H, Jeong J, Kim H, et al. MODi: a powerful and convenient web server for identifying multiple post-translational peptide modifications from tandem mass spectra. Nucleic Acids Res. 2006;34(Web Server issue):W258–63.
Li Q, Shortreed MR, Wenger CD, Frey BL, Schaffer LV, Scalf M, et al. Global post-translational modification discovery. J Proteome Res. 2017;16(4):1383–90.
Yeom J, Kabir MH, Lim B, Ahn HS, Kim SY, Lee C. A proteogenomic approach for protein-level evidence of genomic variants in cancer cells. Sci Rep. 2016;6:35305.
Lawrence RT, Perez EM, Hernandez D, Miller CP, Haas KM, Irie HY, et al. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015;11(4):630–44.
Bertier G, Carrot-Zhang J, Ragoussis V, Joly Y. Integrating precision cancer medicine into healthcare-policy, practice, and research challenges. Genome Med. 2016;8(1):108.
Li L, Wei Y, To C, Zhu CQ, Tong J, Pham NA, et al. Integrated omic analysis of lung cancer reveals metabolism proteome signatures with prognostic impact. Nat Commun. 2014;5:5469.
Huang KL, Li S, Mertins P, Cao S, Gunawardena HP, Ruggles KV, et al. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat Commun. 2017;8:14864.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kang, UB. (2021). Proteomic Interrogation in Cancer Biomarker. In: Noh, DY., Han, W., Toi, M. (eds) Translational Research in Breast Cancer. Advances in Experimental Medicine and Biology, vol 1187. Springer, Singapore. https://doi.org/10.1007/978-981-32-9620-6_15
Download citation
DOI: https://doi.org/10.1007/978-981-32-9620-6_15
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-32-9619-0
Online ISBN: 978-981-32-9620-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)