Abstract
The positive-strand RNA viruses, severe acute respiratory syndrome coronavirus (SARS-CoV) and recently emerged COVID-19 epidemics, demonstrated the transmission capability of the coronaviruses by crossing the species barrier and emergence in humans. The source of coronavirus disease 2019 (COVID-19) is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), firstly reported in December 2019 at Wuhan, China. COVID-19 is a kind of viral pneumonia. The outbreak of SARS-CoV-2 (COVID-19) has been reported as the introduction of the third highly pathogenic coronavirus which crossed the species barrier and spread into the human population. Severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV) were the first two epidemic viruses, respectively, in the twenty-first century. Introduction of the 2019 novel coronaviruses (2019-nCoV) in human population is a worldwide concern, and this might have generated via RNA recombination among the previous reported coronaviruses. The COVID-19 is spreading in an alarming rate, and till date no vaccine or specific medicines are available in the market. The newly emerged coronavirus COVID-19 is strongly related to SARS-CoV except little dissimilarity. In this chapter, we will discuss about the alterations and variations in antigenicity, structural changes, and RNA recombination which might be responsible for the COVID-19 emergence.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
13.1 Introduction
Coronavirus belongs to the large family of viruses, i.e., Coronaviridae family and the order Nidovirales (Cui et al. 2019; Kumar et al. 2020; Phan et al. 2018). Genomic structures and phylogenetic relationship reveals that subfamily Coronavirinae contains the four genera alpha coronavirus and betacoronavirus which are restricted to mammals and responsible for respiratory illness in humans such as Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS coronavirus (SARS-CoV) (Cui et al. 2019; Kumar et al. 2020; Payne 2017; Phan et al. 2018). The other two genera gamma coronavirus and delta coronavirus infect both birds and mammals (Cui et al. 2019; Kumar et al. 2020; Payne 2017; Phan et al. 2018). SARS-CoV and MERS-CoV show severe respiratory diseases in humans, and four others (HCoV-NL63, HCoV-229E, HCoV-OC43, and HKU1) cause normal upper respiratory illness in the hosts which are immunocompetent, even though some can induce severe infections in elders, young children, and infants (Cui et al. 2019; Cascella et al. 2020). Alpha and betacoronaviruses can give rise to intense burden on animals; these include recently emerged swine acute diarrhea syndrome coronavirus (SADS-CoV) and the porcine transmissible gastroenteritis virus and porcine enteric diarrhea virus (PEDV) (Cui et al. 2019; Banerjee et al. 2019).
Continuous development and urbanization increases the frequent integration of many different animals in crowded and populated places may have made easy the emergence and reemergence of a number of these viruses (Lau and Chan 2015; https://www.ncbi.nlm.nih.gov/books/NBK45714/). On the other hand, high mutation and recombination rates are reported in coronaviruses, which may allow the coronaviruses to cross the barrier of species and adopt to new hosts (Lau and Chan 2015; https://clarivate.com/wp-content/uploads/dlm_uploads/2020/01/CORONAVIRUS-REPORT-1.30.2020.pdf; Liu et al. 2020).
The 2003 SARS epidemic has awakened the world’s researchers and scientists on the transmission capability of the coronaviruses from animals to humans. The primary reservoir of the SARS-CoV is horseshoe bat (Lau and Chan 2015; Wang et al. 2006). Current sequence databases revealed the animal origin of the human coronaviruses: MERS-CoV, SARS-CoV, HCoV-229E, and HCoV-NL63 are considered to have originated in bats (Cui et al. 2019; Wang et al. 2006). SARS-associated coronaviruses are found in bats from China and worldwide continuously (Lau and Chan 2015).
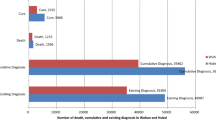
Recently in 2020, SARS-CoV-2, a novel coronavirus has emerged from China which affect globally with a total of 191,127 confirmed cases and 7807 deaths (as of March 18, 2020) (Kampf et al. 2020; https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200314-sitrep-54-covid-19.pdf?sfvrsn=dcd46351_6). Figure 13.1 represents the total reported confirmed cases and deaths due to novel coronavirus (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports). The current pandemic of coronavirus-linked acute respiratory disease known as coronavirus disease 19 (COVID-19) is the third recognized spillover to humans through animal coronavirus in only the last two decades (https://doi.org/10.1038/s41564-020-0695-z; Kampf et al. 2020). Table 13.1 represents the discovery of human coronaviruses.
Total reported confirmed cases and deaths due to novel coronavirus (2019-nCoV). (Data taken from https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports)
The viruses of the family Coronaviridae contain enveloped and positive-stranded RNA (Li et al. 2020; Coutard et al. 2020). Coronaviruses have the largest 26–32 kilobase (kb) positive-sense RNA genome (Li et al. 2020; Kumar et al. 2020; Shoeman and Fielding 2019). The four major structural proteins [membrane (M), envelope (E), spike (S), and nucleocapsid (N)] which create complete virus particle, encoded by the genome (Kumar et al. 2020; Wu et al. 2020; Guo et al. 2020).
The evolution and emergence of novel coronaviruses are mostly due to the lack of proofreading mechanism in RNA recombination among the present coronaviruses (Kumar et al. 2020). The S gene which encodes viral spike (S) glycoprotein has been proposed the higher frequency of recombination (Kumar et al. 2020). The novel coronavirus is closely related to bat SARS like betacoronavirus. Although, the longer spike protein of 2019-nCoV has remarkable difference when compared with the bat SARS-Cov and SARS-like coronaviruses (Kumar et al. 2020). In this chapter, we will discuss about the emergence and reemergence of the SARS coronaviruses and genetic variations from SARS-CoV to SARS-CoV-2.
13.1.1 SARS-CoV to SARS-CoV-2: RNA Recombination, Antigenic Shift, and Antigenic Drift
Reported phylogenetic analysis revealed the close relationship between COVID-19 and Bat SARS-like coronavirus (Lu et al. 2020; Kumar et al. 2020). Although both emerged from the SARS coronavirus, suggesting the newly spread 2019-nCov into human is very much related to SARS-CoV (Kumar et al. 2020). Figure 13.2 represents the evolutionary relationship between different lineage of betacoronavirus and their respective cellular receptors through which they interact with the host cell. As per sequence alignment data, the sequence of spike glycoprotein of COVID-19 and SARS-CoV shows 76.2% individuality, 87.2% resemblance, and 2% gaps. This data indicates that COVID-19 spike glycoprotein exhibits higher sequence similarity with 12.8% of variation with SARS-CoV. The study performed by Kumar et al. (2020), on variation of sequences of minimal receptor-binding domain (RBD), revealed 73.3% uniqueness, 83.9% matches, and 0.4% gaps, suggesting a difference of 16.1% and tertiary structure of minimal receptor-binding domain. COVID-19 may have changes in virus-binding capacity shift and infectivity into the receptors of the host cell, due to notable deviation in minimal RBD of S-glycoprotein.
13.2 Structure of Genome
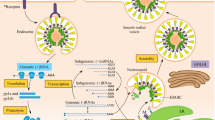
The viruses belong to Coronaviridae family enclosed envelope and positive-stranded RNA (Li et al. 2020). Coronaviruses have the largest 26–32 kilobase (kb) positive-sense RNA genome. The genome of coronaviruses contains open reading frames (ORFs) of variable number (6–11). In the first ORF, two thirds of the total viral RNA are located. Upon infection, two large precursor polyproteins, namely pp1a and pp1ab, has been translated by viral genome in the host cells. These translated precursor polyproteins get processed into 16 mature nonstructural proteins (nsp1–nsp16) by viral proteinases. These nonstructural proteins have been numbered according to the position from N to C terminus (Kumar et al. 2020; Narayanan et al. 2014). Figure 13.3 depicts the structure of coronavirus representing essential proteins, accessory proteins, and nsp1–16. The remaining ORFs encode accessory proteins. The remaining part of viral genome encodes spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid protein (N). All these four are essential structural proteins. S protein is attached to the host receptor ACE2, including two subunits S1 and S2. The cellular tropism by RBD and virus host range is determined by S1. Virus cell membrane fusion is determined by S2 (Guo et al. 2020; Pradhan et al. 2020; Belouzard et al. 2009; Du et al. 2009). Transmembrane nutrient transport and formation of envelop were performed by M protein. N protein and E protein along with several accessory proteins obstruct with host immune response (Guo et al. 2020; Pradhan et al. 2020). The nonstructural proteins play a vital function at the time of viral RNA replication and transcription (Kumar et al. 2020; Narayanan et al. 2014). The assigned functions of these 16 mature nonstructural proteins are mentioned in Table 13.2. Several nsps are unique enzymes implicated in one or more important step(s) in viral replication. Other nsps appear to be entirely involved in virus–host interactions (De Groot et al. 2012).
Coronavirus SARS-CoV-2 is positive ssRNA and enveloped virus. Genome encodes 16 nonstructural proteins (NSPs) and 4 essential structural proteins and several accessory proteins.
13.3 Alteration in Glycosylation Pattern of Spike Glycoproteins
The comparison of the glycosylation sites between COVID-19 and SARS-CoV of the spike glycoproteins unveiled the presence of novel glycosylation sites in COVID-19, such as NGTK, NFTI, NLTT, and NTSN (Kumar et al. 2020). The presence of novel glycosylation sites in COVID-19 might be due to the variation in sequence. Along with the deviation in reported four glycosylation sites, i.e., NGTK, NFTI, NLTT, and NTSN, COVID-19 spike glycoprotein exhibits the similar glycosylation sites, also present in SARS-CoV (NITN, NGTI, NITN, NFSQ, NESL, NCTF, and NNTV). There is a possibility of interaction between COVID-19 and host receptor through novel glycosylation sites which may have an effect on the process of internalization and linked pathogenesis (Kumar et al. 2020).
13.4 Antigenic Variation in Spike Glycoproteins
A study carried out to determine the antigenicity by comparing the antigenic variations in both spike glycoproteins of COVID-19 and SARS-CoV revealed the individuality of most of the CTL epitopes in COVID-19 compared to the SARS-CoV. However, six identical epitopes RISNCVADY, CVADYSVLY, RSFIEDLLF, RVDFCGKGY, MTSCCSCLK, and LKGVKLHY are present in spike glycoprotein of COVID-19 and SARS-CoV (Kumar et al. 2020). In COVID-19, some epitopes are identified with difference in single amino acid change. As per available information and research data of antigenicity, COVID-19 shows little antigenic similarities with SARS coronavirus. It is possible that antigenic similarity may cause similar antigenic response, and therefore, it can be used as one of the precautionary approach. Variation and similarity in spike glycoprotein and epitopes may be useful to design newer and effective vaccines (Kumar et al. 2020).
13.5 Structural Difference in Spike Glycoproteins
The length of encoded proteins in COVID-19 and SARS-like coronaviruses was found nearly same (Guo et al. 2020). However, longer spike protein of COVID-19 showed a notable difference compared to the bat SARS-like coronaviruses and SARS-CoV. On the whole 12.5% of difference in sequences of S glycoprotein and difference in minimal RBD with 23.6% may cause structural differences in spike glycoproteins of COVID-19 and SARS-CoV (Kumar et al. 2020). The studies performed to measure the average distance between the molecules of superimposed proteins, i.e., RMSD, depict the 1.38 Å local RMSD value between two glycoproteins. This illustrates regardless of 12.8% sequence variation that there is an insignificant structural divergence among the spike glycoproteins of SARS-CoV and COVID-19. The absence of structural divergence in spike glycoproteins of SARS-CoV and COVID-19 raises a hope for the treatment of brutal COVID-19. As there is structural similarity in SARS-CoV and COVID-19, the previously trialed and tested attachment inhibitor used for SARS-CoV can be used as the present choice of treatment for COVID-19 (Kumar et al. 2020).
13.6 RNA Recombination in Positive-Strand RNA Viruses
Genetic recombination is an important evolutionary mechanism in positive-strand RNA viruses. Recombination drives toward the diversity in viral genome by the creation of novel chimeric genomes (Bentley and Evans 2018; Loriere and Holmes 2012). Irrespective of single- or multiple-segment genome, RNA recombination may occur in the entire RNA viruses (Loriere and Holmes 2012). Cross-species transmission in RNA viruses is the most common approach to get entered into a new host. The process of recombination assists the entry of viruses as it facilitates viruses to explore a larger proportion of the sequence area, than mutation. This increases the probability of finding a genetic configuration that assists host adaptation (Loriere and Holmes 2012). Both replicative and non-replicative recombination mechanism works for the viruses. In replicative recombination, the major role is played by viral polymerase; however, other viral or cellular proteins may exist. On the contrary, cellular components are solely responsible for non-replicative recombination (Bentley and Evans 2018).
Recently emerged many human diseases are originated from RNA viruses. There are three mechanisms by which viruses go through evolutionary changes. These are mutations also known as antigenic drift, re-assortment (antigenic shift), and recombination (Shao et al. 2017). The emergence of viral diseases in human exhibits active viral genome recombination or re-assortment. Coronavirus that emerged in turkey was a recombinant infectious bronchitis virus which attained a spike protein-encoding gene from coronavirus 122 (Loriere and Holmes 2012). There is a possibility of emergence of COVID-19 through RNA recombination.
Executive Summary
-
The third epidemic of the twenty-first century:
-
SARS-CoV was reported as the first epidemic of the twenty-first century in 2003 following by MERS-CoV in 2012.
-
COVID-19 caused by SARS-CoV-2 is the third epidemic which is declared pandemic by the WHO.
-
-
Severe acute respiratory syndrome (SARS) and coronavirus disease 2019 (COVID-19) portray a number of similarities:
-
Virus homology: As per sequence alignment data, the spike glycoprotein sequence of COVID-19 and SARS-CoV shows 87.2% resemblance; sequences of minimal receptor-binding domain (RBD) show 83.9% similarity.
-
Presence of similar glycosylation sites and similar epitopes.
-
Routes of transmission of both the viruses (i.e., contact, droplets, and fomite).
-
-
Along with the similarities, severe acute respiratory syndrome (SARS) and coronavirus disease 2019 (COVID-19) depict a number of dissimilarities also:
-
Such as 12.5% of difference in S glycoprotein sequence and difference in minimal RBD with 23.6%.
-
Other behavioral differences, such as rate of transmission and severity of COVID-19 is much higher than SARS-CoV.
-
-
The emergence of novel coronavirus 2019 (COVID-19) might be due to RNA recombination as the positive-sense RNA viruses are well reported for their ability of RNA recombination which make them potent to cross the species barrier.
-
The structural similarity of COVID-19 with SARS-CoV could be encashed in terms of:
-
Use of available treatment for the inhibition of attachment with the host cell. This could be an effective way to control the spread of disease.
-
Can use some drug/compound to block the entry into host cell by binding to the receptor.
-
13.7 Conclusions
The newly emerged coronavirus SARS-CoV-2 is strongly related to the predecessor SARS-CoV. SARS-CoV is becoming pandemic which is declared as public health emergency of global concern. The presence of novel glycosylation sites in SARS-CoV-2 makes it possible to become pandemic due to its antigenic divergence. At this time, it is important to understand and educate the mass to follow the instructions given by the authorities and avoid any social gathering. By this at least we can delay the transmission in widespread community. Slowdown of transmission rate will provide time to the researchers and scientists to prepare well and develop vaccine and treatment for this novel coronavirus. The similarity of SARS-CoV-2 to SARS-CoV in terms of antigenic sites proposes the scope of SARS-linked peptide-based vaccine to prevent COVID-19. The structural similarity of SARS-CoV-2 to SARS-CoV suggests the use of attachment inhibitor, specific to coronavirus as the current option of the treatment. Although the mechanism of species to species transfer of the SARS coronavirus is difficult to understand, SARS-CoV 2003 epidemic makes it clear that animal coronaviruses are budding threats to the human community. Still researches are going on COVID-19. Limited information on novel coronavirus makes some boundaries to explain the complete antigenicity and structure of COVID-19.
13.8 Future Perspectives
Complete genome analysis is required to understand the similarities and differences of SARS-CoV-2 with the previously reported viruses. Complete genome analysis also helps in the development of vaccines and medicines against COVID-19. It is required to understand and do the complete research on the emergence and reemergence of coronaviruses and to understand the change in proteins and genome. Transmission through crossing the species barrier is another area to focus on.
Abbreviations
- ACE2:
-
Angiotensin-converting enzyme 2
- DMV:
-
Double membrane vesicle
- HKU:
-
Human coronavirus
- IFN:
-
Interferon
- MERS:
-
Middle East respiratory syndrome
- NSP:
-
Nonstructural protein
- ORF:
-
Open reading frame
- RBD:
-
Receptor-binding domain
- RMSD:
-
Root mean square deviation
- SARS:
-
Severe acute respiratory syndrome
- ssRNA:
-
Single-stranded ribonucleic acid
References
Adnan Shereen M, Khan S, Kazmi A, Bashir N, Siddique R (2020) COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res 24:91–98. https://doi.org/10.1016/j.jare.2020.03.005
Banerjee A, Kulcsar K, Mishra V, Frieman M, Mossman K (2019) Bats and coronaviruses. Viruses 11:41
Belouzard S, Chu VC, Whittaker GR (2009) Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A 106(14):5871–5876
Bentley K, Evans DJ (2018) Mechanisms and consequences of positive-strand RNA virus recombination. J Gen Virol 99:1345–1356
Cascella M, Ranik M, Cuomo A, Dulebohn SC, Napoli RD (2020) Features, evaluation and treatment coronavirus (COVID-19). NCBI Bookshelf: https://www.ncbi.nlm.nih.gov/books/NBK554776/
Chen Y, Liu Q, Guo D (2020) Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol 92:418–423
Clarivate Analytics (2020) Disease briefing: coronaviruses. https://clarivate.com/wp-content/uploads/dlm_uploads/2020/01/CORONAVIRUS-REPORT-1.30.2020.pdf
Coutard B, Valle C, Lamballerie D, Canard B, Seidah NG, DEcroly E (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin like cleavage site absent in CoV of the same clade. Antivir Res 176:104742
Cui J, Li F, Shi ZL (2019) Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192
De Groot RJ, Baker SC, Baric R, Enjuanes L et al (2012) Family—Coronaviridae. Virus taxonomy ninth report of the International Committee on Taxonomy of Viruses, pp 806–828
Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S (2009) The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol 7(3):226–236. https://doi.org/10.1038/nrmicro2090
Guo YR, Cao QD, Hong ZS, Tan YY et al (2020) The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil Med Res 7:11
Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL et al (2011) SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog 7(12):e1002433. https://doi.org/10.1371/journal.ppat.1002433
Hulswit RJG, Langa Y, Bakkersa MJG et al (2019) Human coronaviruses OC43 and HKU1 bind to 9-oacetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci U S A 116(7):2681–2690
Human Coronavirus Types (2020) Centre for Disease control and prevention. https://www.cdc.gov/coronavirus/types.html
Kahn JS (2005) History and recent advances in coronavirus discovery. Pediatr Infect Dis J 24:S223–S227
Kamitani W, Narayanan K, Huang C, Lokugamage K et al (2006) Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A 103(34):12885–12890
Kampf G, Todt D, Pfaender S, Steinmann E (2020) Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J Hosp Infect 104:246–251
Kumar S, Maurya VK, Prasad AK, Bhatt MLB, Saxena SK (2020) Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV). Virus Dis. https://doi.org/10.1007/s13337-020-00571-5
Lau SKP, Chan JFW (2015) Coronaviruses: emerging and re-emerging pathogens in humans and animals. Virol J 12:209. https://doi.org/10.1186/s12985-015-0432-z
Li X, Geng M, Peng Y, Meng L, Lu S (2020) Molecular immune pathogenesis and diagnosis of COVID-19. https://doi.org/10.1016/j.jpha.2020.03.001
Lim YX, Ng YL, Tam JP, Liu DX (2016) Human coronaviruses: a review of virus–host interactions. Diseases 4:26. https://doi.org/10.3390/diseases4030026
Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, Smoot J, Gregg AC, Daniels AD, Jervey S, Albaiu D (2020) Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci 6(3):315–331. https://doi.org/10.1021/acscentsci.0c00272
Loriere ES, Holmes EC (2012) Why do RNA viruses recombine. Nat Rev Microbiol 9(8):617–626. https://doi.org/10.1038/nrmicro2614
Lu R, Zhao X, Li J, Niu P, Yang B et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574
Narayanan K, Ramirez SI, Lokugamage KG, Makino S (2014) Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res 202:89–100. https://doi.org/10.1016/j.virusres.2014.11.019
National Foundation for Infectious Diseases (2020) Coronavirus. https://www.nfid.org/infectious-diseases/coronaviruses/
Parrish CR, Holmes EC, Morens DM, Park EC et al (2008) Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev 72(3):457–470
Payne S (2017) Chapter 17 Coronaviridae. Viruses:149–158. https://doi.org/10.1016/B978-0-12-803109-4.00017-9
Phan MVT, Tri TN, Anh PH, Baker S et al (2018) Identification and characterization of Coronaviridae genomes from Vietnamese bats and rats based on conserved protein domains. Virus Evol 4(2). https://doi.org/10.1093/ve/vey035
Pradhan P, Pandey AK, Mishra A, Gupta P, et al (2020) Uncanny similarity of unique inserts in the 2019-nCoV spike protein to HIV-1 gp120 and Gag. https://doi.org/10.1101/2020.01.30.927871
Rasool SA, Fielding BC (2010) Understanding human coronavirus HCoV-NL63. Open Virol J 2010(4):76–84
Shao W, Li X, Goraya MY, Wang S, Chen JL (2017) Evolution of influenza A virus by mutation and re-assortment. Int J Mol Sci 18:1650
Shoeman D, Fielding BC (2019) Coronavirus envelope protein: current knowledge. Virol J 16:69
Tai W, He L, Zhang X, Pu J et al (2020) Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. https://www.nature.com/articles/s41423-020-0400-4
Wang LF, Shi Z, Zhang S, Field H, Daszak P, Eaton BT (2006) Review of bats and SARS. Emerg Infect Dis 12(12):1834–1840
World Health Organization (2020) Coronavirus disease (COVID-2019) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
Wu C, Liu Y, Yang Y, Zhang P et al (2020) Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. https://doi.org/10.1016/j.apsb.2020.02.008
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Baxi, P., Saxena, S.K. (2020). Emergence and Reemergence of Severe Acute Respiratory Syndrome (SARS) Coronaviruses. In: Saxena, S. (eds) Coronavirus Disease 2019 (COVID-19). Medical Virology: From Pathogenesis to Disease Control. Springer, Singapore. https://doi.org/10.1007/978-981-15-4814-7_13
Download citation
DOI: https://doi.org/10.1007/978-981-15-4814-7_13
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-4813-0
Online ISBN: 978-981-15-4814-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)