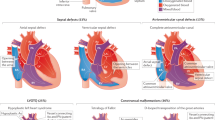
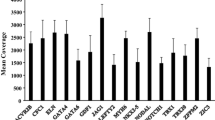
Abstract
Over the last decade, genetic screening has become increasingly available for patients with congenital heart disease (CHD). Chromosomal microarray has proved to be promising, particularly in the diagnosis of syndromic CHDs, and is now considered first-tier cytogenetic testing [1]. Whole-exome sequencing (WES) is another approach for genetic screening aimed towards identifying single-gene mutations [2]. Trio- and pedigree-based WES facilitate the identification of de novo and compound heterozygous variants throughout the entire coding region. Filtering of variants based on allele frequency, in silico predictions and segregation status, effectively narrows down the putative mutation. Thus, application of next-generation sequencing (NGS) has the potential of enhancing gene discovery in CHD pathogenesis.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
Over the last decade, genetic screening has become increasingly available for patients with congenital heart disease (CHD). Chromosomal microarray has proved to be promising, particularly in the diagnosis of syndromic CHDs, and is now considered first-tier cytogenetic testing [1]. Whole-exome sequencing (WES) is another approach for genetic screening aimed towards identifying single-gene mutations [2]. Trio- and pedigree-based WES facilitate the identification of de novo and compound heterozygous variants throughout the entire coding region. Filtering of variants based on allele frequency, in silico predictions and segregation status, effectively narrows down the putative mutation. Thus, application of next-generation sequencing (NGS) has the potential of enhancing gene discovery in CHD pathogenesis.
Despite all the progress, underlying causes for the vast majority of CHDs remain unknown. The present study revealed ~1 de novo coding variant per trio by WES of a heterotaxy cohort. In a familial case of CHD, WES identified compound heterozygous, loss-of-function variants cosegregating with the phenotype [3]. Now, the major challenge lies in interpreting so-called “private” mutations that are specific to each pedigree. Distinguishing a truly causal mutation from benign variants requires well-designed functional studies. Future investigations must focus on establishing vertebrate models that recapitulate human CHD pathogenesis, based on the accumulating mutational data on CHD patients.
References
Nakagama Y, Inuzuka R et al. Chromosomal microarray analysis for the diagnosis of Mowat-Wilson syndrome in two patients with syndromic congenital heart disease. Paper presented at the 52nd annual meeting of the Japanese Society of Pediatric Cardiology and Cardiac Surgery, Tokyo Dome Hotel, Tokyo, 6–8 July 2016.
Nakagama Y, Inuzuka R et al. TGA as a heterotaxy-spectrum heart defect: lessons learned from two primary ciliary dyskinesia cases with AV- or VA-discordant hearts. Paper presented at the 53rd annual meeting of the Japanese Society of Pediatric Cardiology and Cardiac Surgery, Act City Hamamatsu, Shizuoka, 7–9 July 2017.
Nakagama Y, Inuzuka R et al. The role of next-generation sequencing in elucidating the pathogenesis of congenital heart disease. Paper presented at the 8th TAKAO international symposium, Kunibiki Messe, Shimane, 6–8 October 2017.
Acknowledgements
All work was supported by JSPS KAKENHI Grant JP16K10059. Grateful acknowledgement is rendered to Professor Seishi Ogawa, Department of Pathology and Tumor Biology, Graduate School of Medicine, Kyoto University, for support of NGS. The authors also acknowledge the Human Genome Center, Institute of Medical Science, The University of Tokyo, for providing computing resources (https://genomon-project.github.io/GenomonPagesR/).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this paper
Cite this paper
Nakagama, Y., Inuzuka, R. (2020). Elucidating the Pathogenesis of Congenital Heart Disease in the Era of Next-Generation Sequencing. In: Nakanishi, T., Baldwin, H., Fineman, J., Yamagishi, H. (eds) Molecular Mechanism of Congenital Heart Disease and Pulmonary Hypertension. Springer, Singapore. https://doi.org/10.1007/978-981-15-1185-1_65
Download citation
DOI: https://doi.org/10.1007/978-981-15-1185-1_65
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-1184-4
Online ISBN: 978-981-15-1185-1
eBook Packages: MedicineMedicine (R0)