Abstract
Research in the last 10 years has led to improved understanding of the genetic regulation of vertebrate heart development, but despite this effort, approximately 70% of all congenital heart defects (CHDs) still have an unknown etiology. Alternative splicing of mRNA has been documented to play roles in normal and abnormal development. Dysregulated splicing of mRNA has been shown to cause heart defects in mice, however a link between mRNA splicing and CHDs has not yet been shown in humans. We reported that more than 50% of genes associated with heart development were alternatively spliced in the right ventricle (RV) of infants with tetralogy of Fallot (TOF) relative to the RV of normally developing infants. Moreover, there was a significant decrease in the level of 12 scaRNAs (small cajal body associated RNAs) in the RV from infants with TOF. These small noncoding RNAs guide the biochemical modification of specific nucleotides in spliceosomal RNAs that are critical for spliceosomal function. We used primary cells derived from the RV of infants with TOF to show a direct link between scaRNA levels and alteration in mRNA splicing of several genes that regulate heart development. We modified the expression of sets of scaRNAs and consequentially documented distinctive mRNA splicing, accompanied by corresponding protein isoform changes suggesting a unique contribution by each scaRNA. Furthermore, we knocked down two homologous scaRNAs in zebrafish and saw a disruption of heart development with an accompanying alteration in splice isoforms of cardiac regulatory genes. These combined results provide compelling evidence that scaRNAs contribute to the regulation of cardiac development by fine-tuning the fidelity of the spliceosome that adjusts exon retention as cell differentiation occurs. Importantly, our findings are consistent with the concept that disruption of mRNA splicing patterns during early embryonic development disturbs normal signaling pathways, resulting in conotruncal misalignment and TOF.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Congenital heart defects (CHDs) are the most common birth defects and represent a substantial health care burden even in countries with advanced health care systems [1]. Approximately 70% of all CHD cases are idiopathic and a significant effort has been made in the last 10 years to identify the genetic basis of CHDs. However, there remains a significant gap in our understanding of the genetic heritability of CHDs. Splicing in the vertebrate heart has been shown to be dynamic and carefully regulated [2,3,4,5]. However, no direct link between alternative splicing and CHDs has been established. In the developing embryo, precise spatial and temporal signaling is required between the first heart field from which the left ventricle is derived and the second heart field (SHF) from which the right ventricle and the conotruncal outflow tract are derived [3, 6,7,8,9,10,11]. The regulatory networks that control heart morphogenesis are mediated by multiple master regulatory genes (e.g. NKX2.5, GATA4, MBNL1) that control key pathways, including the WNT and NOTCH pathways, all of which have alternatively spliced messages.
Nearly all protein-coding genes in eukaryotic genomes contain introns (94% in humans) which must be removed to produce a mature messenger RNA [12]. Additionally, recent estimates of alternative splicing suggest as many as 90% of human genes have alternative transcript isoforms [13, 14]. There is growing appreciation for the role that alternative splicing plays in normal development and pathogenesis [2, 15,16,17,18,19]. The spliceosome facilitates pre-mRNA processing (splicing) of almost all primary transcripts in eukaryotic genomes. The primary spliceosome, termed the U2 spliceosome, is a multi-megadalton ribonucleoprotein complex composed of numerous proteins and five small nuclear RNAs (snRNAs or spliceosomal RNAs, U1, U2, U4, U5 and U6). The conformation and composition of the spliceosome are highly dynamic and conserved between yeast and humans [20]. Elaborate RNA–RNA–protein interactions align the reactive subgroups and repeatedly rearrange as each intron is identified, intron-exon boundaries are located, and catalysis proceeds to remove each intron in every pre-mRNA.
Some exons are constitutive, that is, they are present in every mature message; however, there are a surprising number of alternatively spliced transcripts that dramatically increase the complexity of the transcriptome and thus the proteome. The significance of alternative splicing for vertebrate evolution was highlighted by two companion papers in the December 21, 2012, issue of Science [21, 22]. Alternative splicing is temporally and spatially controlled resulting in unique splice variants in different tissues and at different time points in the same tissue. Recently, the transition from a fetal to postnatal pattern of a conserved set of alternatively spliced isoforms was shown to regulate mouse heart development [23]. Clearly, mRNA splicing plays a significant role in mammalian cardiac development, but the potential contribution to human heart pathology remains unknown.
Only about 2% of the human genome is translated into protein, but it is now evident that as much as 80% of the genome is transcribed [24]. The importance of noncoding RNA (ncRNA) for heart development has recently been shown by the requirement for correct spatiotemporal expression of specific microRNAs to ensure proper heart development [25]. In addition, there are clear spatial and temporal transcript splicing transitions that are conserved in the vertebrate heart during fetal and postnatal development [23, 26]. It is likely that there are multiple checkpoints to ensure proper transcriptome content for correct heart development. If these checkpoints fail, it is likely that cardiac development will also fail. It is possible that some of the checkpoints may be encoded in ncRNA families yet to be investigated.
A significant class of evolutionarily conserved ncRNA is the small nucleolar RNAs (snoRNAs) with homologs in all eukaryotes. The snoRNAs primarily guide biochemical modifications of specific nucleotides (e.g., methylation and pseudouridylation) of ribosomal RNAs and small nuclear RNAs (snRNAs, also referred to as spliceosomal RNAs). Those snoRNAs that target spliceosomal RNAs are associated with Cajal bodies in the nucleus and are known as scaRNAs (small Cajal-body-specific RNAs). In most vertebrate species, snoRNAs reside in the introns of other genes and require splicing to initiate snoRNA maturation [27]. Interestingly, intron-encoded snoRNAs may have special promoters to drive transcription, suggesting tissue specificity [28]. While the biochemical targets of snoRNAs have been clearly elucidated over the last 20 years, there is a surprising paucity of information regarding the developmental significance of this abundant class of ncRNA.
Our initial studies focused on analyzing the transcriptome of cardiac tissues discarded after surgical correction of tetralogy of Fallot (TOF), a congenital heart disease which is phenotypically representative of disruption of normal conotruncal development. Our preliminary data demonstrated that scaRNAs and their target spliceosomal RNAs are reduced in TOF myocardium [29]. It is therefore possible that reduced levels of scaRNAs may impact stability or fidelity of the spliceosome, causing alterations in mRNA maturation that contributes to TOF. The accumulation of moderate reductions in scaRNA level may cause alterations in spliceosomal function, thus contributing noise to the communication between the first and second heart fields, and resulting in conotruncal misalignment. We hypothesize that tissue-specific control of the pattern of scaRNA expression could provide temporal and spatial specificity to the spliceosome, thus providing a ubiquitous mechanism for regulating splicing in the developing embryo.
While the epigenetic role that scaRNAs play in maintaining spliceosomal integrity and precision is currently unexplored in terms of human health, we have collected substantial evidence that scaRNAs and splicing patterns are tissue-specific and indeed play a critical role in vertebrate cardiac development. We have examined the impact on spliceosome function made by dysregulated scaRNAs using human primary cells derived from the right ventricle of infants with TOF and from normally developing infant heart tissue. In addition, using the well-established zebrafish model, we characterized the nature of scaRNA interaction and its impact on vertebrate heart development. These experiments allowed us to confirm that these epigenetic elements affect vertebrate heart development, which represents a paradigm shift in our understanding of human heart development.
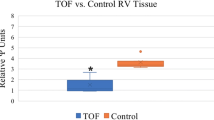
scaRNAs direct the posttranslational biochemical modification of spliceosomal RNAs. Without this modification the spliceosome will fail to function properly [30]. Recent studies show that without site-specific modification the RNA–RNA interactions of the spliceosomal RNAs, U2 and U6 do not achieve the correct conformation [31]. We previously identified 12 scaRNAs that were moderately but significantly reduced in TOF myocardium [29], similar to fetal levels of expression of the scaRNAs (Fig. 53.1). Interestingly, the majority of these scaRNAs target nucleotides that are important for conformational integrity of the U2–U6 complex [31]. Reduced levels of scaRNAs in TOF myocardium suggest a failure of adequate regulation of the scaRNA level early in gestation resulting in inadequate spliceosomal function. In the proposed investigation, we will carefully characterize the synergy between scaRNAs and assess the impact on spliceosome function (i.e., splicing) and vertebrate heart development. It is our contention that scaRNA function may serve as a checkpoint in transcriptional processing, regulating many aspects of development, including heart formation.
2 Subjects
Our subjects were children less than 1 year of age with CHDs: TOF; tranposition of the great arteries (TGA); or pulmonary atresia with intact ventricular septum (PA/IVS) requiring surgical reconstruction. Informed consent was obtained from a parent or legal guardian after reviewing the consent document and having their questions answered (IRB #11120627). Our original observations were based on analysis of tissue from 16 infants with idiopathic TOF (nonsyndromic, without 22q11.2 deletions, 11 males, 5 females), comparison tissues from eight normally developing infants (3 males, 5 females). In addition, we have now characterized scaRNA, spliceosomal RNAs and the splicing pattern of 6 key transcription factors from an additional 21 infants with idiopathic TOF. Our acquisition and characterization of control human infant heart tissues and human fetal heart tissue have been previously described [29, 32]. The fetal hearts were dissected by Dr. James O’Brien, a pediatric cardiac surgeon and co-investigator, who performed the reconstruction of the conotruncal defects to ensure the tissue analyzed was from a similar location as the tissues removed during surgery.
3 Results and Discussion
3.1 scaRNAs Target U2 and U6 snRNAs and Both snRNAs Are Significantly Reduced in TOF
We used the scaRNABase database (http://www-scarna.biotoul.fr/index.php) to identify the nucleotides targeted for modification by the 12 scaRNAs that are reduced in TOF. Only two snRNAs were predicted to be targeted: U2 and U6 snRNAs. Six of the scaRNAs targeted 10 nucleotides (of 23 nucleotides that are known to be modified by scaRNAs) in U2 and 6 scaRNAs targeted 5 nucleotides (of 8 total modified nucleotides) in U6. Interestingly, U2 and U6 had significantly reduced expression in our 16 TOF samples compared to the 8 controls (U2 was reduced 1.8-fold in TOF RV, p = 0.04, and U6 was reduced 3.2-fold in TOF RV p < 0.0001). This is consistent with reduced stability of U2 and U6 as a consequence of inefficient scaRNA biochemical modification.
3.2 Cardiac Regulatory Networks Are Enriched for Alternative Splice Isoforms in TOF
Coordinated control of alternative splicing modifies the transcriptome and hence the proteome, while participating in most developmental processes [33, 34]. Studies of animal models have shown the importance of appropriate splicing for proper heart development [3, 5, 23, 35]. Still, knowledge of the sequence of events leading to tissue-specific alternative splicing is limited. More importantly with respect to the current proposal, nothing is known about the significance of scaRNA-guided nucleotide modification in spliceosomal RNAs and the potential impact on transcript splicing. Intriguingly, our analyses of splicing variants in TOF myocardium revealed a substantial increase of alternative transcript isoforms which were enriched in gene networks known to be critical for regulating heart development, including the WNT and NOTCH pathways. More importantly, ~50% of these alternative isoforms are present in normal fetal RV, suggesting regulation of splicing did not proceed properly during heart development in the infants with TOF (Alternative splicing of mRNA is dynamic in human fetal heart development, in prep). Figure 53.2 shows representative examples of alternative splicing, DICER and DAAM1, which have a similar pattern of splicing in fetal and TOF tissues (blue and green lines) compared to the control tissue (red line). Dicer was shown to be important in mouse heart development playing a role in splicing regulation [3], and splice variants of Daam1, a member of the WNT pathway, were recently shown to impact angiogenesis and endothelial cell migration and tube formation [36].
3.3 Reduced scaRNA Expression and Alternative Splicing Are Not a Consequence of Hypertrophy
Hypertrophy is known to reactivate the expression of some fetal genes. Thus we wanted to determine if the fetal expression patterns we observed could be a consequence of hypertrophy. As a comparison to TOF, we analyzed right ventricular samples from infants with TGA and PA/IVS. The embryological origins of TOF and TGA likely share some common features involving miscommunication between the first and second heart fields. However, PA/IVS is distinct and probably less complex in terms of origin of the regulatory defect. Nevertheless, TOF, TGA and PA/IVS share a common attribute: right ventricular hypertrophy. We observed only ten snoRNAs with differential expression in PA/IVS right ventricular tissue compared to control tissue. The ten snoRNAs that were reduced in PA/IVS were in common with TOF, and all target the 28S rRNA. In addition, U2 and U6 levels were not reduced in PA/IVS relative to the control tissue, and splice isoforms were essentially unchanged relative to the controls. On the other hand, RV from children with TGA bore a striking resemblance to the TOF pattern of scaRNA, spliceosomal RNA expression and fetal type splice isoforms. Therefore, the fetal type pattern of scaRNA and spliceosomal RNA expression, as well as fetal splice isoforms, does not appear to be simply a consequence of hypertrophy since they were not present in PA/IVS RV samples. These findings support our postulation that scaRNAs are of key importance in regulating heart development and not simply a consequence of hypertrophy.
3.4 Primary Cell Lines Derived from TOF Myocardium Retain the Same Relative Expression Patterns as the Tissue
We have derived primary cell lines from right ventricular myocardium obtained from 15 infants with TOF (TOF primary cells—TOFpc). These cells are most likely fibroblasts. The cell type is somewhat inconsequential since what we wish to examine is spliceosome response to changing levels of scaRNAs. We compared scaRNA levels, U2 and U6 levels, and splice isoform patterns between TOFpcs and primary myocytes derived from normally developing infant heart tissue. TOFpcs retained the same fetal type pattern of scaRNA, spliceosomal RNA expression and splicing isoforms of index genes relative to cells derived from normally developing neonatal cardiac tissue (data not shown). Splicing patterns also retained a TOF pattern in the TOFpcs relative to the normal cells. These changes were consistent through at least four passages of the cell lines.
3.5 Overexpression of ACA26 and SCARNA1 (ACA35) in TOF Primary Cells Was Associated with an Increase in U2 Levels and a Decrease in Fetal Splice Isoforms
The scaRNAs were cloned into an intron sequence between hemoglobin exons 3 and 4 so that they would be correctly processed in vivo and expression was driven with the CMV promoter [37]. When either plasmid was transfected alone, there was an increase in the scaRNA but no change in U2 level or in splicing (data not shown). However, when we simultaneously transfected plasmids pCGL-ACA26 and pCGL-SCARNA1 into TOF primary myocytes, there was modest (~50%) but significant upregulation in U2 level. This is consistent with the idea that the scaRNA-directed modification of snRNAs is necessary for snRNA stability. More importantly, there was a reduction in the level of the fetal type splice form (Fig. 53.3). These data were repeated in TOFpcs from three different infants (genotypes). This is an exciting discovery which squarely supports our hypothesis that the scaRNAs are playing a role in spliceosomal function. These novel new findings support the critical role that scaRNAs play in mammalian heart development. This connection has not been investigated previously.
3.6 Knockdown of scaRNAs (scaRNA1 Targeting U2, or snord94 Targeting U6) Causes Heart Defects in Zebrafish
Studies of snoRNA-directed modification of ncRNA in archaea and lower eukaryotes have shown that nucleotide modifications are crucial for ncRNA functions [38, 39]. However, similar studies in vertebrates have not been described until the recent report of the developmental significance of snoRNAs in zebrafish by Kenmochi and colleagues [40]. In summary, they suppressed the expression of several snoRNAs, including U26, in zebrafish (U26 was one of the snoRNAs that we identified as being down regulated in myocardium from infants with TOF). Using a unique highly sensitive mass spec analysis that they developed, they found that decreased U26 snoRNA expression reduced the snoRNA-guided methylation of the target nucleotides. Impaired rRNA modification, even at a single site, led to severe morphological defects and embryonic lethality in zebrafish. Thus, nucleotide modifications in rRNA play an essential role in vertebrate development. Additionally, as a preliminary study this past summer, in collaboration with Kenmochi and colleagues we have suppressed the expression of two scaRNAs, scarna1 and snord94 (both are homologous to scaRNAs identified by our screens of TOF myocardium) in zebrafish embryos. The knockdown of the scaRNAs resulted in developmental abnormalities, including heart malformations (Fig. 53.4) and altered splicing of genes that regulate heart development. This is an exciting finding and taken together with our observations that scaRNA levels impact splicing in TOF primary cardiomyocytes, suggesting that scaRNAs play a critical and, as yet, unrecognized role in vertebrate heart development. Furthermore, it appears that these particular scaRNAs primarily affect heart developmental processes. Collectively, these data provide firm support of our overarching hypothesis.
Representative example of zebrafish morphant at 72 hpf (hours post fertilization, knockdown targeted at scarna1). Arrows indicate the heart, blue and grey outlines indicate enlargement of both the atria and ventricle, respectively. Eye and head size are slightly changed but other organs, brain, notochord, otic vesicles, and fins appear essentially normal at this stage
3.7 Splice Isoforms Change During Development in Zebrafish and After Targeted Knockdown of scaRNAs
We downloaded RNA-Seq data from the Gene Expression Omnibus derived from developing zebrafish at 0.75 h, 6 h, 1 day, 2 days, 3 days and 5 days postfertilization (GEO#: GSE30603) and analyzed for alternative splicing. We found clear changes in ratios of splice isoforms of a large proportion of genes, including genes important for heart development (e.g., Gata4, Mbnl1, Notch1, Dicer, data not shown). We performed RNA-Seq on RNA extracted from 24hpf zebrafish embryos treated with antisense morpholinos directed at scaRNA1 or snord94 and WT untreated embryos and embryos treated with mismatch morpholinos. Paired-end sequencing runs were performed with 101 base reads on the Illumina HiSeq 1500. RNA-Seq data were analyzed using the “Tuxedo suite.” There were no appreciable differences between mismatch morpholino and WT-type embryos. snord94 was reduced by 40%, and scaRNA1 was not detectable in their respective knockdown morphants compared to WT or mismatch treated embryos. We saw a clear shift in the predominant isoforms of our index genes after treatment with the antisense morpholino (Fig. 53.5 shows results of validation of the WNT pathway gene splicing variants in zebrafish morphants). These analyses clearly demonstrate that, as in mammals, splicing in developing zebrafish is dynamic. Furthermore, the knockdown experiments strongly support our hypothesis that scaRNAs are important for regulating splice variants and heart development. Furthermore, snord94 and scaRNA1 appear to be important for heart development.
Splicing of WNT pathway genes is altered in zebrafish morphants. Treating zebrafish embryos with anti-scarna1 morpholino causes changes in exon retention of cardiac regulatory genes. Eleven of 39 members of the WNT family have changes in exon retention after treatment with antisense morpholino (assessed by RNA-Seq and qRT-PCR; values shown are from qRT-PCR data). The mismatch morpholino has no significant effect on splicing
4 Conclusions
We examined the noncoding transcriptome in myocardial tissue from children with tetralogy of Fallot (TOF) and observed changes in mRNA splice isoforms of genes that are critical for regulating heart development [29]. In parallel, we found a modest but significant reduction in the levels of 12 small Cajal body-specific RNAs (scaRNAs are a subset of snoRNAs and direct the biochemical processing of spliceosomal RNA) [29]. These patterns of scaRNA expression and splicing are similar to patterns we saw in the fetal myocardium. To explore the potential relevance of these findings, we manipulated the expression of two of these scaRNAs in cell cultures and zebrafish. We saw clear changes in splicing patterns and, importantly, developmental deficiencies including heart defects in zebrafish. Our findings support a direct role for scaRNAs in fine tuning the fidelity of the spliceosome in a manner that is critical to support splicing transitions that are essential for correct heart development. Our observations open the door on a new paradigm in developmental regulation and potentially may explain a substantial portion of the missing genetic heritability of CHDs. Importantly, by accurately characterizing the role of scaRNAs in disrupting genetic signaling, it is possible that this could lead to the identification of “targets” that may be amenable to change through intervention as is the case with microRNAs.
References
American Heart Association: https://www.heart.org/en/health-topics/congenital-heart-defects (2011).
Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–29.
Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24:653–8.
Bland CS, et al. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res. 2010;38:7651.
Salomonis N, et al. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput Biol. 2009;5:e1000553.
Bentham J, Bhattacharya S. Genetic mechanisms controlling cardiovascular development. Ann N Y Acad Sci. 2008;1123:10–9.
Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–8.
Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–35.
Huang JB, et al. Molecular mechanisms of congenital heart disease. Cardiovasc Pathol. 2009;19:e183–93.
Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–70.
Wessels MW, Willems PJ. Genetic factors in non-syndromic congenital heart malformations. Clin Genet. 2010;78:103–23.
Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220:152–63.
Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470.
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5.
Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472.
Kaida D, Schneider-Poetsch T, Yoshida M. Splicing in oncogenesis and tumor suppression. Cancer Sci. 2012;103:1611–6.
Tanackovic G, et al. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum Mol Genet. 2011;20:2116–30.
Mannoor K, Liao J, Jiang F. Small nucleolar RNAs in cancer. Biochim Biophys Acta. 2012;1826:121–8.
Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12:84–8.
Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3 https://doi.org/10.1101/cshperspect.a003707.
Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–9.
Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–93.
Kalsotra A, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–8.
Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–54.
Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–25.
Castle JC, et al. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–25.
Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–24.
Deng W, et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome Res. 2006;16:20–9.
O’Brien JE Jr, et al. Noncoding RNA expression in myocardium from infants with tetralogy of Fallot. Circ Cardiovasc Genet. 2012;5:279–86.
Karijolich J, Yu YT. Spliceosomal snRNA modifications and their function. RNA Biol. 2010;7:192–204.
Karunatilaka KS, Rueda D. Post-transcriptional modifications modulate conformational dynamics in human U2-U6 snRNA complex. RNA. 2013;20:16–23.
Bittel DC, et al. Gene expression in cardiac tissues from infants with idiopathic conotruncal defects. BMC Med Genomics. 2011;4:1.
Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–4.
Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–61.
Stennard FA, et al. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262:206–24.
Ju R, et al. Activation of the planar cell polarity formin DAAM1 leads to inhibition of endothelial cell proliferation, migration, and angiogenesis. Proc Natl Acad Sci U S A. 2010;107:6906–11.
Darzacq X, et al. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–56.
Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–20.
Liang B, et al. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat Struct Mol Biol. 2009;16:740–6.
Higa-Nakamine S, et al. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res. 2012;40:391–8.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this paper
Cite this paper
Bittel, D.C. et al. (2020). The Role of Alternative mRNA Splicing in Heart Development. In: Nakanishi, T., Baldwin, H., Fineman, J., Yamagishi, H. (eds) Molecular Mechanism of Congenital Heart Disease and Pulmonary Hypertension. Springer, Singapore. https://doi.org/10.1007/978-981-15-1185-1_53
Download citation
DOI: https://doi.org/10.1007/978-981-15-1185-1_53
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-1184-4
Online ISBN: 978-981-15-1185-1
eBook Packages: MedicineMedicine (R0)