Abstract
The bivalve hinge ligament is the hard tissue that functions to open and close shells. The ligament contains fibrous structures consisting of aragonite crystals surrounded by a dense organic matrix. This organic matrix may contribute to the formation of fibrous aragonite crystals, but the mechanism underlying this formation remains unclear. Recently, we showed that tissue inhibitor of metalloproteinase (TIMP) and matrix metalloproteinase (MMP) is related to the formation of the ligament in Pinctada fucata. BLAST search of genome database revealed that seven MMP genes are encoded in the genome of P. fucata. To identify the specific MMP that may contribute to ligament formation, the expression level of each MMP was measured in the mantle isthmus, which secretes the ligament. The expression of MMP54089 increased after scratching of the ligament, while the expressions of other MMPs did not increase after doing the same operation. To identify the role of MMP54089 in forming the ligament structure, double-stranded (ds) RNA targeting MMP54089 was injected into the living P. fucata to suppress the function of MMP54089. Scanning electron microscopic images showed disordered growing surfaces of the ligament in individuals injected with MMP54089-specific dsRNA. These results suggest that PfTIMP and MMP54089 play important roles in the formation of the fibrous ligament structure.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The components of the ligament are secreted by the mantle isthmus, which is the mantle tissue attached to the shell hinge. Bevelander and Nakahara (1969) reported that small fibrous aragonite crystals and organic frameworks are secreted from cells of the mantle isthmus and align to form long aragonite fibers. These fibers and the organic matrix are connected and transported to the shell to form the ligament microstructure. The crystals grow vertically toward the growing surface of the ligament. In a cross section of the ligament, a pseudohexagonal aragonite crystal was observed (Kahler et al. 1976; Marsh and Sass 1980), which appears to be the euhedral shape of the aragonite crystal. The diameter of the crystal is 50–100 nm, and the c axis is parallel to the long axis. In contrast, the a and b axes are oriented randomly. Aragonite crystals with the defect of {110} twinning were observed in the ligament (Kahler et al. 1976; Marsh and Sass 1980).
Suzuki et al. (2015) reported that the aragonite crystals in the ligament of P. fucata contain ligament intracrystalline peptide (LICP). LICP regulates the growth of the aragonite crystal and maintains the orientation of the c axis to keep the aragonite crystals small. These small aragonite crystals can be aligned in the same direction and stacked to form aragonite fibers. The mechanism by which these small crystals are gathered and arranged in the organic matrix of the ligament is still unknown. To reveal the mechanisms underlying the formation of the complicated aragonite microstructures present in the ligament, structural and functional analyses of the organic matrix between the aragonite fibers are necessary. Recently, we reported that tissue inhibitor of metalloproteinase (TIMP) was identified from the insoluble fraction of the ligament in P. fucata (Kubota et al. 2017). This report shows the function of matrix metalloproteinase (MMP) for the formation of the ligament.
2 Materials and Methods
We used the BLAST search method to identify the MMP genes. We also prepared three types of pearl oyster samples to synthesize the cDNA for the templates of RT-PCR: sample 1 received no wound as a control, sample 2 received a wound on the shell by breaking the edge of the outer side of the shell with a nipper as the non-specific injured control, and sample 3 received a wound on the ligament from the outside to the inside with a cutter. After making a wound, each oyster was cultured in about 10 L seawater for 36 h. The PCR used the following cycling conditions: 35 cycles of 30 s at 94 °C (3 min 30 s for the first cycle), 30 s at 55 °C, and 30 s at 72 °C. The expression level of each MMP was checked by agarose gel electrophoresis of the PCR products. The gel contained 0.004% ethidium bromide so that amplified DNA could be visualized. After the electrophoresis, the fluorescence of the gel was observed under the exposure of the light at the wavelength of 365 nm.
dsRNAs of MMP genes were prepared for the RNAi experiments. 30 μg of each dsRNA was dissolved with 50 μL PBS buffer and injected to adductor muscle of Pinctada fucata. We used four young oysters (7 cm in shell size) in this experiment, because it was necessary to observe the growth surface of the ligament. Four days after injection, the growth surface of the ligament was observed by scanning electron microscope (SEM). We observed the ligament growth edges of three individuals using SEM independently. These observations showed similar results. Table 39.1 shows the list of primers used in these experiments.
3 Results
3.1 MMP Genes in P. fucata
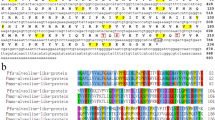
Blast search showed seven MMP genes in the P. fucata genomic database (Takeuchi et al. 2012): pfu_aug1.0_21.1_21914 (21914), pfu_aug1.0_1282.1_07860 (07860), pfu_aug1.0_11733.1_60936 (60936), pfu_aug1.0_14122.1_32404 (32404), pfu_aug1.0_4795.1_23659 (23659), pfu_aug1.0_14629.1_54089 (54089), and pfu_aug1.0_322.1_14973 (14973) (Fig. 39.1). Among these MMPs, some conserved domains were identified. All MMPs in the database have the MMP superfamily domain that contains the activity site of MMP. MMP21924, MMP07862, MMP32404, MMP54089, and MMP14973 have a putative peptidoglycan-binding domain. No putative peptidoglycan-binding domain is found in human MMPs. MMP21914, MMP07860, MMP23659, and MMP14973 have hemopexin-like repeat domain, which is also found in most of human MMPs. Only MMP14973 has a signal peptide in the N-terminal sequence.
The domain structures of MMPs from P. fucata. MMP21914 consists of a putative peptidoglycan-binding domain, an MMP superfamily domain, and hemopexin-like repeats. MMP07860 consists of a putative peptidoglycan-binding domain, an MMP superfamily domain, and hemopexin-like repeats. MMP60936 consists of only an MMP superfamily domain. MMP32404 contains of a putative peptidoglycan-binding domain and an MMP superfamily domain. MMP23659 consists of an MMP superfamily domain and hemopexin-like repeats. MMP54089 consists of a putative peptidoglycan-binding domain and an MMP superfamily domain. MMP14973 consists of a putative peptidoglycan-binding domain, an MMP superfamily domain, and hemopexin-like repeats. Black boxes show the putative peptidoglycan-binding domain. Dotted boxes show the hemopexin-like repeats. Slashed boxes show the MMP superfamily domain. Gray box shows the signal peptide
3.2 MMP Gene Expressions in Response to Wounds
To further investigate the expression level of each MMP in the mantle isthmus, the wound repair process was investigated, and MMP expression levels during this process were compared (Fig. 39.2). We assumed that expression of the MMP related to formation of the ligament would increase during the wound repair response of the ligament. Living pearl oysters were cultured in an aquarium for 1 week at 20 °C after the shell had been damaged by a knife. We used three treatment conditions: a control without any wounds, a wound on the shell margin, and a wound on the ligament. Significant changes in the expression levels of MMPs 32404, 23659, and 54089 were detected under these three conditions. Although the expression of MMP32404 decreased when the oyster was wounded on its shell margin, the expression levels in the control was higher than that in the ligament wound treatments, indicating that MMP32404 did not play a role in regeneration of the ligament. The expression level of MMP23659 increased after both shell margin and ligament wounds, indicating that MMP23659 has a non-specific wound-repair function in the shell. The expression level of MMP54089 increased only in the ligament wound treatment, indicating that MMP54089 plays a role specifically in regeneration of the ligament.
3.3 RNAi Experiment
To investigate the function of MMP54089 in the ligament, an RNAi experiment was performed. dsRNA targeting MMP54089 was injected into four living pearl oysters, which were then cultured in an aquarium for 4 days at 20 °C. The hinge ligament is attached to the nacreous layer of the shell (Fig. 39.3a, b). The growing edge of the ligament was observed by SEM 4 days after injection. In the control treatment of (EGFP)-specific dsRNA injection, the tips of aragonite crystals were observed as white dots along the surface of the growing edge (Fig. 39.3c). Similar microstructures were observed after injections with MMP14973 dsRNA (Fig. 39.3d). In contrast, disordered microstructure was observed after MMP54089-specific dsRNA injection (Fig. 39.3e). The disordered organic matrix exhibited a wavy surface, and the aragonite crystals were not aligned, suggesting that MMP54089 plays a key role in the regulated formation of the fibrous microstructure of the ligament.
(a) The shell of P. fucata. The white rectangle showed the region observed in (b). (b) SEM observations of the ligament and nacreous layer. (c) SEM observations of the surface in the ligament’s growing edge in the sample of EGFP-dsRNA injection. (d) The surface in the ligament’s growing edge in the sample of MMP14973-dsRNA injection. (e) The surface in the ligament’s growing edge in the sample of MMP54089-dsRNA injection
4 Discussion
Two hypotheses regarding the functions of PfTIMP and MMP54089 in the ligament were formulated. First, MMP54089 may degrade extracellular organic fibers to soften them and increase the space between them. The loosened extracellular organic fibers tend to adjust to the same orientation. A previous report showed that physical stress increased the activity of human MMP2 as well as its expression and also increased the toughness of a blood vessel analog in a cell-seeded collagen gel (Seliktar et al. 2003). A similar phenomenon may occur in the ligament of P. fucata. MMP54089 of P. fucata degrades extracellular organic fibers in the ligament and arranges them to regulate the direction of their growth. In the second hypothesis, MMP54089 digests proteins to prepare the peptides that contribute to calcium carbonate crystallization. A previous work suggested that amelogenin in the enamel of human teeth is digested by human MMP20, and the peptide fragments of amelogenin promote the crystallization of calcium phosphorus in the teeth (Vuk et al. 2011). MMP54089 may digest some matrix proteins such as LICP to prepare the released peptides for calcium carbonate crystallization (Suzuki et al. 2015). Further investigation is required to reveal the relationships among PfTIMP, MMP54089, extracellular organic fibers, and calcium carbonate crystallization.
References
Bevelander G, Nakahara H (1969) An electron microscope study of the formation of the ligament of Mytilus edulis and Pinctada radiata Calcif. Tiss Res 4:101–112
Kahler G, Sass R, Fisher F (1976) The fine structure and crystallography of the hinge ligament of Spisula solidissima (Mollusca: Bivalvia: Mactridae). J Comp Physiol 109:209–220
Kubota K, Tsuchihashi Y, Kogure T, Maeyama K, Hattori F, Kinoshit S, Sakuda S, Nagasawa H, Yoshimura E, Suzuki M (2017) Structural and functional analyses of a TIMP and MMP in the ligament of Pinctada fucata. J Struct Biol 199:216–224
Marsh M, Sass R (1980) Aragonite twinning in the molluscan bivalve hinge ligament. Science 208:1262–1263
Seliktar D, Nerem R, Galis Z (2003) Mechanical strain-stimulated remodeling of tissue-engineered blood vessel constructs. Tissue Eng 9:657–666
Suzuki M, Kogure T, Sakuda S, Nagasawa H (2015) Identification of ligament intra-crystalline peptide (LICP) from the hinge ligament of the bivalve, Pinctada Fucata. Mar Biotechnol 17:153–161
Takeuchi T, Kawashima T, Koyanagi R, Gyoja F, Tanaka M, Ikuta T, Shoguchi E, Fujiwara M, Shinzato C, Hisata K, Fujie M, Usami T, Nagai K, Maeyama K, Okamoto K, Aoki H, Ishikawa T, Masaoka T, Fujiwara A, Endo K, Endo H, Nagasawa H, Kinoshita S, Asakawa S, Watabe S, Satoh N (2012) Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res 2:117–130
Vuk U, Feroz K, Haichuan L, Halina E, Li Z, Wu L, Stefan H (2011) Hydrolysis of amelogenin by matrix metalloproteinase-20 accelerates mineralization in vitro. Arch Oral Biol 56:1548–1559
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2018 The Author(s)
About this paper
Cite this paper
Kubota, K. et al. (2018). Functional Analyses of MMP Genes in the Ligament of Pinctada fucata . In: Endo, K., Kogure, T., Nagasawa, H. (eds) Biomineralization. Springer, Singapore. https://doi.org/10.1007/978-981-13-1002-7_39
Download citation
DOI: https://doi.org/10.1007/978-981-13-1002-7_39
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1001-0
Online ISBN: 978-981-13-1002-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)