Abstract
In barnacle shell, the calcified shell layer is laid on top of the epicuticle. Here, we report our strategy and some preliminary results on the identification of potential shell proteins of the barnacle Megabalanus rosa. At first, M. rosa proteins from acid-soluble and acid-insoluble shell extracts were subjected to proteomic analysis and searched against M. rosa complete transcriptome. Then using the information that the calcified shell is formed just after the larval-adult molt, juvenile differentially expressed genes against larval stages were screened. Sixty secretory protein sequences were identified as primary candidates of M. rosa shell proteins, among which 37 are novel proteins.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Thoracican barnacles produce a heavily calcified shell that, unlike their crustacean relatives, does not shed away (Walley 1969). The calcified shell offers physical protection against predators and wave actions (Astachov et al. 2011). The shell also enables barnacles to prevent desiccation in the intertidal region.
The calcified shell plates produced by the subtidal thoracican barnacle Megabalanus rosa are composed of the operculum plates (OPs), the lateral wall plates (WPs), and the basal shell plate (BP) (Fig. 11.1). OPs consist of two pairs of shell plates, namely, terga and scuta. These are movable plates that serve as the shutter of the soft body. M. rosa produces four pairs of WPs, which are the major shell plates that protect the interior soft body from mechanical impact. BP is the calcified shell plate at the bottom of the barnacle, separating the barnacle soft body from the rock substratum surface. Hence, each of these plates has different functions. Details of barnacle shell anatomy can be found in Chan et al. (2009).
The thoracican barnacle Megabalanus rosa and a schemetic drawing of the adult shell structure. (a) 2-month M. rosa juvenile, with cirri reaching out from its calcified shell for feeding on planktons. (b) A schemetic drawing of M. rosa shell structure. M. rosa shell composes of the operculum plates (OPs), the wall plates (WPs), and a basal plate (BP)
Obviously, it is important to understand what molecules are involved in the formation of these shell structures. Matsubara et al. (2008) isolated two C-type lectin proteins BRA-2 and BRA-3 from the regenerating M. rosa shell. Zhang et al. (2015) reported a list of acid-soluble shell proteins, including carbonic anhydrase and an aspartic acid-rich partial protein sequence from the intertidal barnacle Amphibalanus amphitrite. The aspartic acid-rich partial protein, however, has shown no sequence homology to the molluscan Asprich shell protein family (Gotliv et al. 2005), suggesting that aspartic acid-rich proteins in barnacles and mollusks have independently evolved.
In this study, we used M. rosa as the study model for barnacle shell formation. We present an integrated transcriptomic and proteomic approach for the identification of barnacle shell protein candidates.
2 Material and Methods
2.1 M. rosa and Its Larval Culture Construction
Adult individuals of M. rosa were obtained in the Oga Peninsula, Akita, Japan, from the bouy deployed by local fishermen. Upon arriving the lab, the M. rosa individuals were maintained in a 20 °C aquarium until being sampled. Larval samples were prepared at Central Research Institute of Electric Power Industry, according to Yoshimura et al. (2006a, b).
2.2 Transcriptome Data Analysis
The details of the preparation of M. rosa Illumina transcriptome will be reported elsewhere. The M. rosa transcriptome assembly covered various larval stages, attached larvae, metamorphosing larvae, 1-day and 3-day-old juveniles, and adult tissues. The transcriptome data were mainly assembled using Trinity v2.3.2 (Grabherr et al. 2011). But we soon discovered that Trinity failed to resolve highly repetitive tandem repeat structures often found among potential shell protein transcripts, owing to the use of fixed short kmer length (kmer = 25 bp). Therefore, we switched to a multiple kmer strategy, in which the Illumina read data were assembled using Transabyss v1.6.3 (Robertson et al. 2010) with kmer size ranging from 21 to 161 bp, for every 10 bp. These assemblies were then merged using the script Transabyss-merge. Transcript abundance estimation was performed using the software RSEM (Li and Dewey 2011). The levels of transcript expression were normalized according to coverage and then converted to the relative expression levels in FPKM values. Differential expression analysis was performed using the software edgeR (Robinson et al. 2010).
2.3 Preparation of Shell-Soluble and Shell-Insoluble Fractions
To extract shell proteins, the mineralized shell plates, namely, OPs, WPs, and BP as described above, were subjected to acid decalcification. After removing the promosa (all soft body parts) from the shell, OPs, WPs, and BP were then cleaned by 10% bleach solution (household bleach) for 3 days, with daily exchange of the bleach solution. After bleaching, the shell plates were washed with distilled water extensively and then dried in freeze drier for 1 day. The dried shell plates were then crushed into fine powder smaller than 110 μm (filtered by a mesh filter). Altogether, 20 M. rosa specimens (integument diameter ranging from 2 to 5 cm) were being sacrificed. Then, around 12 g of OP powder, 20 g of WP powder, and 20 g of BP powder were independently decalcified by 10% acetic acid overnight in 4 °C. By 4000 × g centrifugation, the supernatants, which represent the acid-soluble fractions (ASF), and the pellets, which represent the acid-insoluble fractions (AIF) of OP, WP, and BP, were separated. Acetic acid in the ASF of OP, WP, and BP was exchanged with Milli-Q water by dialysis (6–8 kDa dialysis membrane). Proteins in the ASFs were then concentrated by 3 kDa cutoff centrifugal filtering device (Amicon Ultra, Millipore). After removal of lipids by methanol-chloroform precipitation, purified ASF protein samples were redissolved in 40 μl 2 M thiourea. To extract proteins from AIF, Milli-Q water-washed insoluble pellets from OP, WP, and BP were treated with 1 ml of 8 M urea for 1 h at room temperature. The retrieved solubilized AIFs were then subjected to methanol-chloroform precipitation. The purified AIF proteins were redissolved in 40 μl 2 M thiourea. ASF and AIF protein samples were then loaded onto a 4–20% SDS-PAGE gradient gel. The gel was subsequently stained with Coomassie Brilliant Blue G-250.
2.4 In-Gel Digestion, LC-MS/MS Analysis, and Protein Identification
The AIF and ASF samples from OPs, WPs, and BP were divided into nine size fractions and independently retrieved for in-gel digestion. Destaining, reduction, alkylation, in-gel digestion, and the final peptide digest retrieval were performed as in Sun et al. (2012), except that 1 μg of trypsin (sequencing grade) dissolved in 100 μl of 10 mM of triethylammonium bicarbonate was applied to the gel pieces from each sample; standard shotgun proteomics techniques were used to analyze the samples on a Thermo Scientific LTQ VelosTM platform (Thermo Fisher Scientific, Bremen, Germany). Operation details of the LC-MS/MS can refer to Zhang et al. (2014). Identification of protein was performed using MASCOT search engine (v2.3.02), with the parameter setting the same as in Zhang et al. (2014). The transcriptome data generated from M. rosa (method and sample details mentioned above) were in silico translated to a protein database using Transdecoder v.3.0.3 (Haas and Papanicolaou 2016), with minimum length of the open reading frame set at 100 amino acid. Decoy sequences consisting of shuffled protein sequences for each of the proteins were concatenated to the original protein database to generate a target-decoy database for protein identification.
2.5 Analysis of M. rosa Shell Protein Candidates
The identified protein sequences were analyzed for the presence of N-terminal signal sequence using SignalP server 4.1 (Nielsen 2017), followed by manual correction. The identified proteins with N-terminal signal peptide were then selected and categorized based on their annotation status.
3 Results and Discussion
3.1 Development of Shell Formation in M. rosa Juvenile
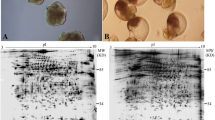
Figure 11.2a shows the life cycle of thoracican barnacles including M. rosa. M. rosa larvae are in general encased in chitinous cuticles. They do not produce any calcified shell structure at any larval stage. Formation of calcium carbonate shell occurs only after post-settlement metamorphosis, that is, the transition of a cypris larva into a juvenile barnacle. This means that the genes responsible for barnacle shell formation start to be expressed during metamorphosis and in the early juvenile stage. As shown in Fig. 11.2b, in M. rosa, the progression of calcification was the most obvious at the first 5 days of juvenile development. At 5 days after metamorphosis, all shell plates are completely nontransparent, indicating these shell plates are all mineralized.
Life cycle of M. rosa and the development of young juvenile. (a) The barnacle spawned as nauplius larva, which undergoes four successive molts and then transforms into a cypris larva. The cypris larva actively searches for an appropriate substratum and will then fully commit to settlement, which involves attachment of the larval body onto the substratum by cement secretion, and metamorphosis, which involves ecdysis of the cyprid body molt. The soft newly metamorphosed juvenile soon begins to feed on planktons using the cirri and develops into an adult barnacle, protected by calcified shells. (b) Development of young juvenile. The 0 day juvenile, that is, the stage when ecdysis was just completed. Five days after completion of metamorphosis, the juvenile has developed mineralized shell plates
3.2 Identification of Proteins in M. rosa Shell Extract
In total, 431 translated protein sequences (Fig. 11.3a) from all the 6 samples, that is, the AIF and ASF of BP/OPs/WPs, were identified using the translated protein database generated from the multiple kmer Transabyss pipeline (contains 35,998 translated nonredundant protein sequences).
Proteins identified from the acid-soluble and acid-insoluble extract of all the shell parts of M. rosa. (a) The whole shell proteome as identified in this study. Four hundred and thirty-one translated protein sequence identified by Mascot protein identification pipeline. (b) Shell proteome filtered by transcriptome data. Sixty translated proteins with a predicted N-terminal signal peptide and whose transcripts were 16 times upregulated during metamorphosis and the juvenile stages
3.3 Filtering Shell Proteome Data with Transcriptome Analysis
We noticed that, among the 431 identified translated protein sequences identified from all the shell samples, ovary proteins such as vitellogenin were detected (Fig. 11.3a), indicating that the shell plate samples were contaminated by ovary tissue, and bleaching obviously was not effective in removing all of these ovary proteins stuck to the shell during sampling. From our preliminary SEM and EDX analyses, we have observed that at the juvenile stage, shell material is being actively synthesized. We therefore selected the identified proteins whose transcripts were four-folds (16 times) more highly expressed in the juvenile stages than in the larval stages (FDR <0.001). This criterion filtered out 349 proteins, leaving 82 relevant shell proteins, among which 60 contain a signal peptide (Fig. 11.3b). With the juvenile-specific expression pattern and the presence of signal peptide as the criteria, the list of identified shell protein candidates became much smaller yet specific. Among the 60 shell protein candidates, some of them were implicated in shell formation in other systems, including carbonic anhydrase, C-type lectin domain protein, and chitin-binding domain proteins (Miyamoto et al. 1996; Suzuki et al. 2007; Joubert et al. 2010). In addition, 37 unknown/novel proteins were also discovered, and these proteins are now under our major investigation. Our results clearly indicate that our transcriptome-based filtering method, that is, to filter the shell proteome data with the corresponding transcript expression patterns, is an excellent pipeline for the purpose of relevant shell protein identification.
References
Astachov L, Nevo Z, Brosh T, Vago R (2011) The structural, compositional and mechanical features of the calcite shell of the barnacle Tetraclita rufotincta. J Struct Biol 175:311–318
Chan BKK, Prabowo RE, Lee KS (2009) Crustacean fauna of Taiwan: barnacles, volume I-Cirripedia: thoracica excluding the Pyrgomatidae and Acastinae. National Taiwan Ocean University, Keelung
Gotliv BA, Kessler N, Sumerel JL, Morse DE, Tuross N, Addadi L, Weiner S (2005) Asprich: a novel aspartic acid-rich protein family from the prismatic shell matrix of the bivalve Atrina rigida. ChemBioChem 6:304–314
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Haas BJ, Papanicolaou A (2016) TransDecoder (finding coding regions within transcripts)
Joubert C, Piquemal D, Marie B, Manchon L, Pierrat F, Zanella-Cléon I, Cochennec-Laureau N, Gueguen Y, Montagnani C (2010) Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: focus on biomineralization. BMC Genomics 11:613
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323
Matsubara H, Hayashi T, Ogawa T, Muramoto K, Jimbo M, Kamiya H (2008) Modulating effect of acorn barnacle C-type lectins on the crystallization of calcium carbonate. Fish Sci 74:418–424
Miyamoto H, Miyashita T, Okushima M, Nakano S, Morita T, Matsushiro A (1996) A carbonic anhydrase from the nacreous layer in oyster pearls. Proc Natl Acad Sci U S A 93:9657–9660
Nielsen H (2017) Predicting secretory proteins with SignalP. In: Protein function prediction: methods and protocols. Springer, New York, pp 59–73
Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, Griffith M (2010) De novo assembly and analysis of RNA-seq data. Nat Methods 7:909–912
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Sun J, Zhang H, Wang H, Heras H, Dreon MS, Ituarte S et al (2012) First proteome of the egg perivitelline fluid of a freshwater gastropod with aerial oviposition. J Proteome Res 11(8):4240–4248
Suzuki M, Sakuda S, Nagasawa H (2007) Identification of chitin in the prismatic layer of the shell and a chitin synthase gene from the Japanese pearl oyster, Pinctada fucata. Biosci Biotechnol Biochem 71:1735–1744
Walley LJ (1969) Studies on the larval structure and metamorphosis of Balanus balanoides (L.). Philos Trans R Soc B 256:237–280
Yoshimura E, Nogata Y, Sakaguchi I (2006a) Experiments on rearing the barnacle Megabalanus rosa to the cyprid stage in the laboratory. Sessile Organisms 23(2):33–37
Yoshimura E, Nogata Y, Sakaguchi I (2006b) Simple methods for mass culture of barnacle larvae. Sessile Org 23:39–42
Zhang Y, Sun J, Mu H, Li J, Zhang Y, Xu F et al (2014) Proteomic basis of stress responses in the gills of the pacific oyster Crassostrea gigas. J Proteome Res 14(1):304–317
Zhang G, He LS, Wong YH, Xu Y, Zhang Y, Qian PY (2015) Chemical component and proteomic study of the Amphibalanus (= Balanus) amphitrite shell. PLoS One 10(7):e0133866
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2018 The Author(s)
About this paper
Cite this paper
Wong, Y.H. et al. (2018). Identification of Barnacle Shell Proteins by Transcriptome and Proteomic Approaches. In: Endo, K., Kogure, T., Nagasawa, H. (eds) Biomineralization. Springer, Singapore. https://doi.org/10.1007/978-981-13-1002-7_11
Download citation
DOI: https://doi.org/10.1007/978-981-13-1002-7_11
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1001-0
Online ISBN: 978-981-13-1002-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)