Abstract
Information of genetic diversity is very important in supporting the implementation of genetic resource conservation and plant breeding. This study aims to determine the genetic diversity of 11 sago palm accessions that have been collected by the Sago Research Consortium (SRC), University of Papua (UNIPA). Methods of research were divided into three steps. Step 1 was DNA extraction by using Geneaid DNA plant Mini Kit. Step 2 was PCR amplification fragments DNA by using KAPA2G Robust HotStart polymerases and performed by using Bio-Rad PCR instrument. Step 3 was purified and sequenced DNA by Macrogen Inc. Seoul, Korea. Data analyses were performed by using a MEGA6.06 software. Morphological characteristics in the rosette stage of 11 sago palm accessions were divided into 4 phenotypes only, and several accessions showed the same phenotypes. Based on atp6-2 gene markers, the 11 sago palm accessions had a different genetic characteristic among the others. Genetic distances among 11 sago palm accessions were in the range among 0.22 and 3.01 with an average 1.21 pairwise distant. Phylogenetic construction showed that the genetic relationship of 11 sago palm accessions was clustered into 6 groups. The accession numbers SP001 and SP002 are in group 1, the accession numbers SP003 and SP011 group 2, the accession numbers SP005 and SP007 group 3, the accession number SP009 group 4, the accession numbers SP008 and SP010 group 5, and the accession numbers SP004 and SP006 group 6. Nucleotide sequence alignments of mitochondrial atp6-2 gene and introns were different from each other in 11 accessions.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Indonesia has the largest area of sago plantings and sago forests, as well as the richest genetic diversity of the palm. World sago plantations and sago forests were estimated at 2 million ha, with 1 million ha located in Indonesia (Flach 1983). Kertopermono (1996) reported a more extensive area of sago palm in Indonesia totaling 1,528,917 ha, with 1,406,469 in Papua, 45,540 in Sulawesi, 41,949 in Maluku, 31,872 in Sumatra, 2795 in Kalimantan, and 292 ha in West Java. Sago palms in Indonesia are unevenly distributed, as is their genetic diversity. Flach (1983) estimated the diversity of sago as being higher in Papua than elsewhere in the country.
Plant characters such as morphology, protein, and deoxyribonucleic acid (DNA) can be used as markers to ascertain the diversity of sago palms. Morphology and protein characters are insufficient as indicators to reveal genetic diversity because they are influenced by the environment. DNA markers are not influenced by the environment and therefore are used to establish genetic diversity. Determining the genetic diversity of sago palm populations in Indonesia has focused on diversity based on DNA markers of the nuclear, chloroplast, and mitochondrial genomes. DNA markers used for the analysis are as follows: random amplified polymorphism DNA (RAPD), the expression of specific genes (waxy gene) in the nucleus genome, chloroplast simple sequence repeat (cpSSR) in the chloroplast genome, and the expression of functional genes (gene atp6) in the mitochondrial genome.
2 Genetic Diversity of Sago Palm Based on the Nuclear Genome
2.1 RAPD Markers
A total of 100 sago palm samples were collected from 6 locations (Papua, Maluku, Sulawesi, Kalimantan, Java, and Sumatra) and 9 populations (Jayapura, Serui, Manokwari, Sorong, Maluku, Palopo, Pontianak, Bogor, and Selat Panjang) of sago palm centers in several parts of Indonesia.
Polymorphisms of RAPD amplification fragments using 10 RAPD primers and performed with PCR tools resulted in 86 polymorphic fragments and 2–7 genotype numbers per population (Abbas et al. 2009). High numbers of RAPD polymorphisms and genotypes were found in this observation. These results were similar to the genetic diversity of sago palm found in an earlier study by Ehara et al. (2003), which used RAPD markers employing a small number of individual sago palm samples from Indonesia and Malaysia.
Numbers of fragmented DNA bands were amplified from each primer and exhibited a range of 6–12 polymorphic bands per primer; no monomorphic DNA band was observed. The average number of polymorphic DNA bands was calculated to be 9 per primer. Primer P17 resulted in the highest numbers of polymorphic DNA bands produced, 12, whereas primers OPA04 and P06 produced the lowest numbers of polymorphic DNA bands, 6 per primer. The size of DNA bands produced by 10 primer sets had a range of 150–1800 bp (base pairs). Overall primers in this analysis were suitable to study sago palm genetics (Abbas et al. 2009). A previous observation of this involved assaying more than 100 RAPD primers sets.
The genetic diversity of sago palm populations from six Indonesia locations is presented in Table 5.1. The average number of individuals (RIP) of each population is 2–18, the number of polymorphic sites (S) is 16–86, the mean pairwise differences (π) is 10.8333–30.6457, and genotype diversity (Ĥ) equals 1. Variant diversity of genotype (VĤ) is 0.0026–0.5000, and the percentage of polymorphic loci (P) is 18.6047–98.8372. The highest values of S, π, and P are in Papua, meaning that the sago palms there are the most diverse. The H value of all samples overall in Papua is 1.000, meaning all samples are different. The highest VĤ value is from Java (Table 5.1) because samples from that location were the fewest in number and different from each other.
2.2 Waxy Gene Markers
A total of a 100 samples of sago palm were collected from Jayapura, Serui, Manokwari, Sorong, Maluku, Palopo, Pontianak, Bogor, and Selat Panjang populations of sago palm in Indonesia. A total of 8 polymorphic alleles and 14 genotypes of Wx genes were observed in the sago palm genome. Alleles ranging from 100 to 700 bp were detected on agarose gels (Abbas and Ehara 2012). Polymorphism levels detected in sago palm were similar to those found in the wheat (Triticum aestivum L.) genome by Shariflou and Sharp (1999), using a Wx gene (Sun1) primer. Also, Wx gene variations observed in the sago palm genome were similar to those found by Boggini et al. (2001) in the wheat genome. Using single nucleotide genotyping of the Wx gene, high polymorphism in barley was reported by Domon et al. (2004) as well as in rice by Bao et al. (2002) and Tran et al. (2011), using the microsatellite marker for Wx genes.
The distribution of genotypes of sago palm showed that the populations possessing a specific genotype are found in Serui in Papua and in the population of Palopo in Sulawesi. This indicates that both Papua and Sulawesi are the origin of sago palm variation because of the existence of specific genotypes in their populations; both locations are, therefore, considered to be the original sources of sago palm germplasm in Indonesia. Papua exhibited the largest genotype numbers existing in their population based on Wx gene markers; therefore, it is considered to be the center of sago palm genetic diversity in Indonesia.
The genetic diversity of sago palm samples from several areas in Indonesia was calculated. The values of S, G, Ĥ, VĤ, π, and P, based on 8 alleles and 14 genotypes of Wx gene markers in a total of 100 sago palm samples, all showed variations, except in the samples from Java (Table 5.2). The samples reflect sago palm variations in several of the locations. The Wx gene is one of the genes active in the starch biosynthesis process, especially for synthesizing amylose in plants such as the sago palm, which produce starch. Variations of Wx genes as found in the sago palm genome indicate variations in the quality and quantity of starch produced. Ehara et al. (2005) reported that sago palm stems produced 28–710 kg of starch; Yamamoto et al. (2005) reported that starch production per trunk varied among varieties. Local people experienced in working with sago palms report that the quality of starch varied in terms of color and the quality of pappeda, a type of porridge made from it.
The quality and the quantity of starch may be affected by the genes that regulate the starch biosynthesis process, among them the Wx gene. Larkin and Park (2003) found that the Wx gene is one of the many genes that play a part in starch synthesis in the endosperm of rice. Furthermore, Wanchana et al. (2003) found two alleles of the Wx gene (Wxa and Wxb) that regulated the enhancement of the protein and amylose contents of the starch. The lack of a Wx allele in wheat significantly decreased amylose production (Sharma et al. 2002). Epstein et al. (2002) reported that recombinant inbred lines (RILs) of wheat, which have three Wx genes in their genome, produced better-quality starch in comparison with wheat RILs which do not contain three Wx genes.
3 Genetic Diversity of Sago Palm Based on the Chloroplast Genome
Polymorphism of cpDNA
Primers rpl1671, NTCP21, and NTCP22 resulted in PCR products of 100 samples. No polymorphisms were detected on three of the agarose gels, but SNF analysis using fluorescence primers showed polymorphisms on 97 of the samples. The nucleotide detections clearly appeared in each individual sample. Primer pairs of rpl1671-FAM produced four alleles in the size range 147–406 bp, primer pairs of NTCP21-HEX produced five alleles in the size range of 76–406 bp, and primer pairs of NTCP22-NED produced two alleles in the range of 75–160 bp. In a previous study, primer pairs of NTCP21 and NTCP22 also detected polymorphism alleles in potato (Bryan et al. 1999).
Haplotype Identification and Composition
In the three cpDNA loci (rpl1671, NTCP21, and NTCP22) of sago palm, there were observed 10 haplotypes and 11 alleles in the populations of sago palm (data not shown). Haplotypes H01 and H02 were located in two cpDNA loci, and haplotypes H03–H10 were located in three cpDNA loci. Haplotypes H01, H02, H07, and H09 were found spread into two or more populations (Abbas et al. 2010). Only small individual sago palms were moved from a certain population to others because only four of ten haplotypes were shared in two or more populations, which indicated that only four haplotypes of sago palm were estimated to migrate in many ways. In related studies using cpDNA, markers were also reported in refugial populations of Scots pine (Pinus sylvestris L.) and silver fir (Abies alba Mill.) (Provan et al. 1998; Vendramin et al. 1999).
Haplotype H05 was found only in the population from Palopo, Sulawesi, with a 0.25 frequency, and haplotype H10 was found only in the Jayapura population of Pontianak, Kalimantan, with a 0.06 frequency. Haplotypes H04, H06, and H08 were only found in the Jayapura population with 0.04, 0.04, and 0.12 frequencies, respectively. Haplotype H03 was found only in the Serui population with a 0.04 frequency, so that we found four specific haplotypes in Papua (Abbas et al. 2010). In these observations, we found six specific haplotypes which were distributed in Papua, Sulawesi, and Kalimantan. Specific haplotype phenomena have also found in alyssum flowers (Alyssum spp.) by Mengoni et al. (2003) and in cypress (Cunninghamia spp.) by Hwang et al. (2003). Specific haplotypes within populations indicate that they should be the origin of diversity, but we cannot state that the populations of sago palm are diverse because they also have a haplotype with a shared overall population (H07).
Sago palm populations in Papua (Jayapura and Serui), Sulawesi (Palopo), and Kalimantan (Pontianak) which possess specific haplotypes could be the origin of sago palm diversity. The magnitude of haplotype numbers can be used as one of several indicators to indicate the center of diversity. Vendramin et al. (1999) and Mengoni et al. (2003) reported that high numbers of haplotypes indicated a high level of variation. The presence of a widespread common haplotype indicates a major ancient population (Provan et al. 2001). Papua could be the origin and center of sago palm diversity because it has the highest specific haplotypes and the highest numbers of haplotypes, as well as the existence of wild types. Haplotypes described in this study must have existed in the distant past because cpDNA markers exhibit highly conservative sequences (Provan et al. 2001), very low mutation rates which range from 3.2 × 10−5 to 7.9 × 10−5 (Provan et al. 1999), and no recombinant DNA (Ishikawa et al. 1992; Provan et al. 2001) and are uniparentally inherited (Savolainen et al. 1995; Viard et al. 2001). The highest numbers of haplotypes indicate the highest variation as occurred in populations of Abies alba Mill. (Vendramin et al. 1999). Mengoni et al. (2003) documented that the differentiation of chloroplast haplotypes in a population reflected genetic entity. Other reports were similar to our observation; Flach (1983, 1997) documented that Papua had the highest sago palm diversity based on morphological characters and the largest wild stands. Based on the diversity and extensive wild stands, Papua should be considered as the center of diversity (Flach 1997). In Papua, the highest sago palm variety based on morphological characters and environmental adaptability is recognized (Yamamoto et al. 2005).
Genetic Diversity
The genetic diversity of sago palm within Indonesia shows that Papua has the highest values of haplotype numbers, polymorphic sites numbers, and percentages of haplotype polymorphisms. Java has the highest mean pairwise differences (Table 5.3). Haplotype diversity values (HE) among individuals were relatively higher than the overall populations. The HE value of 1 indicates that no haplotype sharing was found in the sample (single haplotype) or samples were different from the others such as occurred in Java. These features probably occurred because the sample sizes were very limited and the sampling criteria in the population were based on phenotypic differences. In a previous study of sago palm using RAPD markers, a high variance in individuals and a high level of sago palm diversity were observed by Ehara et al. (2003) and were also observed using AFLP by Celiz et al. (2004). In related cases, gene pool variations were also recorded in individual Scots pines (Pinus sylvestris L.) within woodlands (Provan et al. 1998).
4 Genetic Diversity of Sago Palm Based on the Mitochondrial Genome
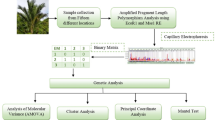
Sago palms used in this study were derived from several regions in Papua with the materials collected by the Sago Research Consortium, University of Papua (SRC, UNIPA). DNA extraction followed the procedures of the Plant Genomic DNA Mini Kit (Geneaid 2012). The design of atp6-2 primer sequences was based on Duminil et al. (2002), and the primers were synthesized by the Genetika Science Company. The primer sequences used for analyses of mitochondrial genetic diversity were forward 5′ GCA TCA TTC AAG TAA ATA CA 3′ and reverse 5′ GTG AAG CTG TCT GGA GGG 3′.
Variation of mtDNA
Amplification of mitochondrial DNA (mtDNA) by using a primer pair of mitochondrial atp6-2 genes and their introns from the 11 accessions sago palm was successfully carried out. The sizes of the nucleotide sequences of mt atp6-2 genes and their introns were detected in the accessions SP001, SP002, and SP003 to SP011 in 829, 825, 772, 651, 855, 640, 680, 690, 629, 728, and 651 bp, respectively. The mt atp6-2 gene and their introns from the 11 sago palm accession genomes had high polymorphism with many different nucleotides (Abbas et al. 2015). The phenomenon observed in 11 sago palm accessions resembles those of pine trees which showed a high level of diversity in the mitochondrial genome (Wu et al. 1998). The high levels of nucleotide diversity of mt atp6-2 genes in the 11 sago palm accessions indicate that they were all different. Touzet and Delph (2009) reported that gynodioecious plants have high nucleotide diversity and form many haplotypes; otherwise hermaphrodite and dioecious species have low nucleotide diversity. Furthermore, Uwai et al. (2006) reported intraspecies variation in the mitochondrial cox3 genes of the seaweed Undaria pinnatifida.
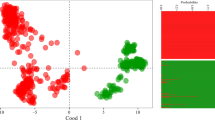
Genetic distances of sago palm accessions were calculated at 0.22–3.01 with an average of 1.21. The 11 sago palm accessions showed a significant difference in the level of genetic distance (data not showed). Phylogenetic construction to demonstrate the genetic relationships of the 11 sago palm accessions showed that the accessions were clustered into six groups. Group 1 consists of accession numbers SP001 and SP002; group 2, SP003 and SP011; group 3, SP005 and SP007; group 4, SP009; group 5, SP008 and SP010; and group 6, SP004 and SP006 (Fig. 5.1). Groups are based on the level of mitochondrial nucleotide polymorphisms of the 11 accessions, in particular genes that encode atp6-2 and their introns. Accessions which have a highly similar genetic value formed the same group in an adjacent phylogenetic branch. Differentiation hypothetically occurred over a long time period because mt markers are maternally inherited and have a low mutation rate. Plant mitochondrial genomes possess a low mutation rate, little compactness, large size, and highly rearranged structure (Darracq et al. 2011). Furthermore, it was reported that the mt genomes of plants have a mechanism of base excision repair pathway (Boesch et al. 2009) so that the nucleotide structure is very conservative. Phylogenetic construction of the 11 sago palm accessions was relatively inclined to form many groups because the high level of mt atp6-2 gene polymorphisms. High polymorphisms of the atp6-2 gene in sago palm may be caused by silent segregation. That phenomenon resembles the mitochondrial atp6 genes which occur in bladder campion, Silene vulgaris (Barr et al. 2007). Furthermore, Barr et al. (2007) also reported that the polymorphism levels of certain genes were variable in plant mitochondrial genomes.
5 Conclusions
Based on the observations, facts, and analytical review of the genetic diversity of sago palm resources in Indonesia, the following conclusions can be made:
-
1.
Molecular markers of sago palm resources in Indonesia show a high variation based on the RAPD polymorphism level and genetic diversity calculations.
-
2.
Wx genes in sago palm are related to starch production variation. Variations of Wx genes in sago palm show a high polymorphism level, with 14 genotypes, high heterozygous value, and high diversity.
-
3.
Sago palm resources in Indonesia are variable based on cpDNA markers, with ten haplotypes found along with high diversity.
-
4.
Based on atp6-2 gene markers, the sago palm accessions showed different genetic characteristics.
-
5.
Sago palm dispersal in Indonesia was not thwarted by distance and geographic isolation.
-
6.
Vernacular names of sago palms given by local people were not related to genetic characters based on RAPD, Wx genes in the nucleolus genome, cpDNA in the chloroplast genome, and atp6-2 in the mitochondrial genome, neither within a population nor among populations.
-
7.
The center of sago palm diversity in Indonesia is proposed as Papua, while the source of diversity is proposed in addition in Sulawesi and Kalimantan, also based on haplotype distribution of cpDNA markers.
References
Abbas B, Ehara H (2012) Assessment genetic variation and relationship of sago palm (Metroxylon sagu Rottb.) in Indonesia based on specific expression gene (Wx genes) markers. African J Plant Sci 6(12):314–320
Abbas B, Bintoro MH, Sudarsono S et al (2009) Genetic relationship of sago palm (Metroxylon sagu Rottb.) in Indonesia based on RAPD markers. J Biol Div 10(4):168–174
Abbas B, Renwarin Y, Bintoro MH et al (2010) Genetic diversity of sago palm in Indonesia based on chloroplast DNA (cpDNA) markers. J Biol Div 11(3):112–117
Abbas B, Listyorini FH, Munarti (2015) Genetic diversity of eleven sago palm accessions fromSRC’s germ plasm based on mitochondrial atp6-2 genes and introns. Paper presented at 12th International Sago Symposium, Tokyo
Bao JS, Corke H, Sun M (2002) Microsatellites in starch-synthesizing genes in relation to starch physicochemical properties in waxy rice (Oriza sativa L.) Theor Appl Genet 105:898–905
Barr CM, Keller SR, Ingvarsson PK et al (2007) Variation in mutation rate and polymorphism among mitochondrial gene of Silene vulgaris. Mol Biol Evol 24(8):1783–1791
Boesch P, Ibrahim N, Paulus F et al (2009) Plant mitochondria possess a short-patch base excision DNA repair pathway. Nucleic Acids Res 37(17):5690–5700
Boggini G, Cttaneo M, Paganoni P, Vaccino P (2001) Genetic variation for waxy proteins and starch properties in Italian wheat germplasm. Euphytica 119:111–114
Bryan GJ, McNicol J, Ramsay G et al (1999) Polymorphic simple sequence repeat markers in chloroplast genomes of solanaceous plants. Theor Appl Genet 99:859–867
Celiz LL, Toyota K, Okazaki M, Power A (2004) Genetic characteristic of sago palm (Metroxylon sagu) cultivars using AFLP (amplified fragment length polymorphism) markers. Proceeding of Japanese Society Sago Palm, pp 61–65
Darracq A, Varre JS, Drouard LM et al (2011) Structural and content diversity of mitochondrial genome in beet: a comparative genomic analysis. Genom Biol Evol 3:723–736
Domon E, Yanagisawa T, Saito A, Takeda K (2004) Single nucleotide polymorphism genotyping of the barley Waxy gene by polymerase chain reaction with confronting two-pair primers. Plant Breed 123(3):225–228
Duminil J, Pomonge MH, Petit RJ (2002) A set of 35 consensus primer pairs amplifying genes and introns of plant mitochondrial DNA. Mol Ecol Notes 2:428–430
Ehara H, Kosaka S, Shimura N et al (2003) Relationship between geographical distribution and genetic distance of sago palm in Malay Archipelago. Sago Palm 11:8–13
Ehara H, Naito H, Mizota C (2005) Environment factors limiting sago production and genetic variation in Metroxylon sagu Rottb. Eighth International Sago Symposium in Jayapura, Indonesia. Japan Society for the Promotion Science, pp 93–104
Epstein J, Morris CF, Huber KC (2002) Instrumental texture of white salted noodles prepared from recombinant inbred lines of wheat differing in the three granule bound starch synthase (waxy) genes. J Cereal Sci 35(1):51–63
Flach M (1983) The sago palm: domestication, exploitation, and product. FAO Plant Production and Protection, Rome
Flach M (1997) Sago palm Metroxylon sagu Rottb. Promoting the conservation and use of underutilized and neglected crops. IPGRI, Rome
Geneaid (2012) Genomic DNA mini kit (plant) protocol. http://www.geneaid.com/sites/default/files/GP9.pdf (Acces July 3, 2013)
Hwang SY, Lin TP, Ma CS et al (2003) Postglacial population growth of Cunninghamia konishii (Cupressaceae) inferred from phylogeographical and mismatch analysis of chloroplast DNA variation. Mol Ecol 12:2689–2695
Ishikawa S, Kato S, Imakawa S et al (1992) Organelle DNA polymorphism in apple cultivars and rootstocks. Theor Appl Genet:963–967
Kertopermono AP (1996) Inventory and evaluation of sago palm (Metroxylon sp.) distribution. Sixth International Sago Symposium. Pekan Baru 9–12 Dec 1996, pp 59–68
Larkin PD, Park WD (2003) Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oriza sativa L.) Mol Breed 12(4):335–339
Mengoni A, Gonelli C, Brocchini C et al (2003) Chloroplast genetic diversity and biogeography in the serpentine endemic Ni-hyperaccumulator Alyssum bertolonii. New Phytol 157:349–356
Provan J, Soranzo N, Wilson NJ et al (1998) Gene-pool variation in Caledonian and European Scots pine (Pinus sylvestris L.) revealed by chloroplast simple-sequence repeats. Proc Biol Sci 265(1407):1697–1705
Provan J, Soranzo N, Wilson NJ et al (1999) A low mutation rate for chloroplast microsatellites. Genetics 153:943–947
Provan J, Powel W, Hollingsworth M (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16(3):142–147
Savolainen V, Corbaz R, Moncousin C et al (1995) Chloroplast DNA variation and parentage analysis in 55 apples. Theor Appl Genet 90:1138–1141
Shariflou MR, Sharp PJ (1999) A polymorphic microsatellite in the 3′ end waxy genes of wheat, Triticum aestivum. Plant Breed 118:275–277
Sharma R, Sison MJ, Rathjen AJ, Jenner CF (2002) The null allele at the waxy locus in durum wheat affect pasta cooking quality. J Cereal Sci 35(3):287–297
Touzet P, Delph LF (2009) The effect of breeding system on polymorphism in mitochondrial genes of sine. Genetics 181:631–664
Tran NA, Daygon VD, Resurreccion AP et al (2011) A single nucleotide polymorphism in the waxy gene explains a significant component of gel consistency. Theor Appl Genet 123:519–525
Uwai S, Yotsukura N, Serisawa Y et al (2006) Intraspecific genetic diversity of Undaria pinnatifida in Japan, based on the mitochondrial cox3 gene and the ITS1 of nrDNA. Hydrobiology 553:345–356
Vendramin GG, Degen B, Petit RJ (1999) High level of variation at Abies alba chloroplast microsatellite loci in Europe. Mol Ecol 8:1117–1126
Viard F, Kassaby YAE, Ritland K (2001) Diversity and genetic structure in populations of Pseudotsuga menziesii (Pinaceae) at chloroplast microsatellite loci. Genome 44:336–344
Wanchana S, Toojinda T, Tragoonrung S, Vanavichit A (2003) Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.) Plant Sci 16(6):1193–1199
Wu J, Krutovskii KV, Strauss SH (1998) Abundant mitochondrial genome diversity, population differentiation, and convergent evolution in pines. Genetics 159:1605–1614
Yamamoto Y, Yoshida T, Miyazaki A et al (2005) Biodiversity and productivity of several sago palm varieties in Indonesia. Eighth International Sago Symposium in Jayapura, Indonesia. Japan Society for the Promotion Science, pp 35–40
Acknowledgments
I am grateful to the management of the MP3EI project with SPPK No. 162/SP211/LT/DRPM/III/2016 from DRPM Directorate General of Strengthen Research and Development (DGSRD) Indonesia for providing financial support in fiscal year 2016. Thanks also to all who contributed to and reviewed this work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
This chapter is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
Copyright information
© 2018 The Author(s)
About this chapter
Cite this chapter
Abbas, B. (2018). Sago Palm Genetic Resource Diversity in Indonesia. In: Ehara, H., Toyoda, Y., Johnson, D. (eds) Sago Palm. Springer, Singapore. https://doi.org/10.1007/978-981-10-5269-9_5
Download citation
DOI: https://doi.org/10.1007/978-981-10-5269-9_5
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-5268-2
Online ISBN: 978-981-10-5269-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)