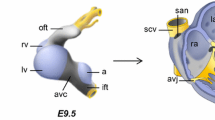
Abstract
Cardiac development is a fine-tuned process governed by complex transcriptional networks, in which transcription factors (TFs) interact with other regulatory layers. In this chapter, we first introduce the core cardiac TFs including Gata, Hand, Nkx2, Mef2 and Tbx, and Srf. These factors regulate each other’s expression and also can act in a combinatorial manner on their downstream targets. Their disruption leads to various cardiac phenotypes in mice, and mutations in humans have been associated with congenital heart defects. In the second part of the chapter, we discuss different levels of regulation including cis-regulatory elements, chromatin structure, and micro-RNAs, which can interact with transcription factors, modulate their function, or are downstream targets. Finally, examples of disturbances of the cardiac regulatory network leading to congenital heart diseases in human are provided.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Buckingham M, Meilhac S, Zaffran S (2005) Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 6:826–835
Cai C-L, Liang X, Shi Y et al (2003) Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell 5:877–889
Engleka KA, Manderfield LJ, Brust RD et al (2012) Islet1 derivatives in the heart are of both neural crest and second heart field origin. Circ Res 110:922–926
Kathiriya IS, Nora EP, Bruneau BG (2015) Investigating the transcriptional control of cardiovascular development. Circ Res 116:700–714
Toenjes M, Schueler M, Hammer S et al (2008) Prediction of cardiac transcription networks based on molecular data and complex clinical phenotypes. Mol Biosyst 4:589–598
Schlesinger J, Schueler M, Grunert M et al (2011) The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet 7, e1001313
He A, Kong SW, Ma Q, Pu WT (2011) Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A 108:5632–5637
Visel A, Blow MJ, Li Z et al (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457:854–858
Lage K, Møllgård K, Greenway S et al (2010) Dissecting spatio-temporal protein networks driving human heart development and related disorders. Mol Syst Biol 6:381
Takeuchi JK, Bruneau BG (2009) Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459:708–711
Ieda M, Fu J-D, Delgado-Olguín P et al (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142:375–386
McCulley DJ, Black BL (2012) Transcription factor pathways and congenital heart disease. Curr Top Dev Biol 100:253–277
Kamohara H, Sakamoto K, Ishiko T et al (1994) Human carcinoma cell lines produce biologically active leukemia inhibitory factor (LIF). Res Commun Mol Pathol Pharmacol 85:131–140
Takeuchi JK, Ohgi M, Koshiba-Takeuchi K et al (2003) Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development 130:5953–5964
Yamagishi H, Yamagishi C, Nakagawa O et al (2001) The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev Biol 239:190–203
Vincentz JW, Barnes RM, Firulli BA et al (2008) Cooperative interaction of Nkx2.5 and Mef2c transcription factors during heart development. Dev Dyn 237:3809–3819
Sepulveda JL, Vlahopoulos S, Iyer D et al (2002) Combinatorial expression of GATA4, Nkx2-5, and serum response factor directs early cardiac gene activity. J Biol Chem 277:25775–25782
Lyons I, Parsons LM, Hartley L et al (1995) Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev 9:1654–1666
Jay PY, Harris BS, Maguire CT et al (2004) Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest 113:1130–1137
Clark KL, Yutzey KE, Benson DW (2006) Transcription factors and congenital heart defects. Annu Rev Physiol 68:97–121
Benson DW (2010) Genetic origins of pediatric heart disease. Pediatr Cardiol 31:422–429
Potthoff MJ, Olson EN (2007) MEF2: a central regulator of diverse developmental programs. Development 134:4131–4140
Iida K, Hidaka K, Takeuchi M et al (1999) Expression of MEF2 genes during human cardiac development. Tohoku J Exp Med 187:15–23
Amacher SL, Buskin JN, Hauschka SD (1993) Multiple regulatory elements contribute differentially to muscle creatine kinase enhancer activity in skeletal and cardiac muscle. Mol Cell Biol 13:2753–2764
Lange M, Kaynak B, Forster UB et al (2008) Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev 22:2370–2384
Ghosh TK, Song FF, Packham EA et al (2009) Physical interaction between TBX5 and MEF2C is required for early heart development. Mol Cell Biol 29:2205–2218
Olson EN (2006) Gene regulatory networks in the evolution and development of the heart. Science 313:1922–1927
Karamboulas C, Dakubo GD, Liu J et al (2006) Disruption of MEF2 activity in cardiomyoblasts inhibits cardiomyogenesis. J Cell Sci 119:4315–4321
Lu J, McKinsey TA, Zhang CL, Olson EN (2000) Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell 6:233–244
Chen J-F, Mandel EM, Thomson JM et al (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38:228–233
Naya FJ, Black BL, Wu H et al (2002) Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med 8:1303–1309
Wang L, Fan C, Topol SE et al (2003) Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 302:1578–1581
Lin Q, Schwarz J, Bucana C, Olson EN (1997) Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276:1404–1407
Verzi MP, McCulley DJ, De Val S et al (2005) The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol 287:134–145
Nemer M (2008) Genetic insights into normal and abnormal heart development. Cardiovasc Pathol 17:48–54
Plageman TF, Yutzey KE (2005) T-box genes and heart development: putting the “T” in heart. Dev Dyn 232:11–20
Mesbah K, Rana MS, Francou A et al (2012) Identification of a Tbx1/Tbx2/Tbx3 genetic pathway governing pharyngeal and arterial pole morphogenesis. Hum Mol Genet 21:1217–1229
Hatcher CJ, Basson CT (2009) Specification of the cardiac conduction system by transcription factors. Circ Res 105:620–630
Bruneau BG, Nemer G, Schmitt JP et al (2001) A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106:709–721
Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K et al (2005) Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development 132:2463–2474
Maitra M, Schluterman MK, Nichols HA et al (2009) Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol 326:368–377
Basson CT, Bachinsky DR, Lin RC et al (1997) Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 15:30–35
Smemo S, Campos LC, Moskowitz IP et al (2012) Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum Mol Genet 21:3255–3263
Merscher S, Funke B, Epstein JA et al (2001) TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104:619–629
Meneghini V, Odent S, Platonova N et al (2006) Novel TBX3 mutation data in families with ulnar-mammary syndrome indicate a genotype-phenotype relationship: mutations that do not disrupt the T-domain are associated with less severe limb defects. Eur J Med Genet 49:151–158
Kirk EP, Sunde M, Costa MW et al (2007) Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet 81:280–291
Ma L, Li J, Liu Y et al (2013) Novel and functional variants within the TBX18 gene promoter in ventricular septal defects. Mol Cell Biochem 382:121–126
Peterkin T, Gibson A, Loose M, Patient R (2005) The roles of GATA-4, -5 and -6 in vertebrate heart development. Semin Cell Dev Biol 16:83–94
Pikkarainen S, Tokola H, Kerkelä R, Ruskoaho H (2004) GATA transcription factors in the developing and adult heart. Cardiovasc Res 63:196–207
Dai Y-S, Cserjesi P, Markham BE, Molkentin JD (2002) The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J Biol Chem 277:24390–24398
Lee Y, Shioi T, Kasahara H et al (1998) The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol 18:3120–3129
Morin S, Charron F, Robitaille L, Nemer M (2000) GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J 19:2046–2055
Belaguli NS, Sepulveda JL, Nigam V et al (2000) Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol Cell Biol 20:7550–7558
Garg V, Kathiriya IS, Barnes R et al (2003) GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424:443–447
Moorman AFM, Christoffels VM (2003) Cardiac chamber formation: development, genes, and evolution. Physiol Rev 83:1223–1267
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705
Davis FJ, Gupta M, Camoretti-Mercado B et al (2003) Calcium/calmodulin-dependent protein kinase activates serum response factor transcription activity by its dissociation from histone deacetylase, HDAC4. Implications in cardiac muscle gene regulation during hypertrophy. J Biol Chem 278:20047–20058
He A, Gu F, Hu Y et al (2014) Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat Commun 5:4907
Molkentin JD, Lin Q, Duncan SA, Olson EN (1997) Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev 11:1061–1072
Pu WT, Ishiwata T, Juraszek AL et al (2004) GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol 275:235–244
Srivastava D, Thomas T, Lin Q et al (1997) Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet 16:154–160
Tsuchihashi T, Maeda J, Shin CH et al (2011) Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev Biol 351:62–69
Riley PR, Gertsenstein M, Dawson K, Cross JC (2000) Early exclusion of hand1-deficient cells from distinct regions of the left ventricular myocardium in chimeric mouse embryos. Dev Biol 227:156–168
McFadden DG, Barbosa AC, Richardson JA et al (2005) The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132:189–201
Dodou E, Verzi MP, Anderson JP et al (2004) Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development 131:3931–3942
Laugwitz K-L, Moretti A, Caron L et al (2008) Islet1 cardiovascular progenitors: a single source for heart lineages? Development 135:193–205
Stevens KN, Hakonarson H, Kim CE et al (2009) Common variation in ISL1 confers genetic susceptibility for human congenital heart disease. PLoS One 5:e10855
Miano JM, Long X, Fujiwara K (2007) Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol 292:C70–C81
Parlakian A, Tuil D, Hamard G et al (2004) Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol Cell Biol 24:5281–5289
Belaguli NS, Schildmeyer LA, Schwartz RJ (1997) Organization and myogenic restricted expression of the murine serum response factor gene. A role for autoregulation. J Biol Chem 272:18222–18231
Wang D, Chang PS, Wang Z et al (2001) Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105:851–862
Cao D, Wang Z, Zhang C-L et al (2005) Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol Cell Biol 25:364–376
Schueler M, Zhang Q, Schlesinger J et al (2012) Dynamics of Srf, p300 and histone modifications during cardiac maturation in mouse. Mol Biosyst 8:495–503
Kook H, Lepore JJ, Gitler AD et al (2003) Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest 112:863–871
Cordes KR, Sheehy NT, White MP et al (2009) miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460:705–710
Chen L, Fulcoli FG, Tang S, Baldini A (2009) Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ Res 105:842–851
ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA et al (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799–816
Ernst J, Kellis M (2013) Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types. Genome Res 23:1142–1154
Narlikar L, Sakabe NJ, Blanski AA et al (2010) Genome-wide discovery of human heart enhancers. Genome Res 20:381–392
Guo Z, Maki M, Ding R et al (2014) Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci Rep 4:5150
Kaynak B, von Heydebreck A, Mebus S et al (2003) Genome-wide array analysis of normal and malformed human hearts. Circulation 107:2467–2474
Zaidi S, Choi M, Wakimoto H et al (2013) De novo mutations in histone-modifying genes in congenital heart disease. Nature 498:220–223
Grunert M, Dorn C, Schueler M et al (2014) Rare and private variations in neural crest, apoptosis and sacromere genes define the polygenic background of isolated Tetralogy of Fallot. Hum Mol Genet 23:3115–3128
Greenway SC, Pereira AC, Lin JC et al (2009) De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet 41:931–935
Theodoris CV, Li M, White MP et al (2015) Human disease modeling reveals integrated transcriptional and epigenetic mechanisms of NOTCH1 haploinsufficiency. Cell 160:1072–1086
Sotoodehnia N, Isaacs A, de Bakker PIW et al (2010) Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet 42:1068–1076
van den Boogaard M, Wong LYE, Tessadori F et al (2012) Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest 122:2519–2530
Acknowledgements
This work was supported by the European Community’s Seventh Framework Programme contract (“CardioNeT”) grant 289600 to SRS and the German Research Foundation (Heisenberg professorship and grant 574157 to SRS). This work was also supported by the Berlin Institute of Health (BIH-CRG2-ConDi to SRS).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer-Verlag Wien
About this chapter
Cite this chapter
Grunert, M., Dorn, C., Rickert-Sperling, S. (2016). Cardiac Transcription Factors and Regulatory Networks. In: Rickert-Sperling, S., Kelly, R., Driscoll, D. (eds) Congenital Heart Diseases: The Broken Heart. Springer, Vienna. https://doi.org/10.1007/978-3-7091-1883-2_12
Download citation
DOI: https://doi.org/10.1007/978-3-7091-1883-2_12
Publisher Name: Springer, Vienna
Print ISBN: 978-3-7091-1882-5
Online ISBN: 978-3-7091-1883-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)