Abstract
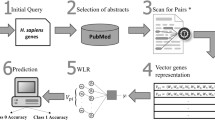
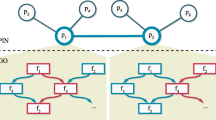
With the extraordinarily increase in genomic sequence data, there is a need to develop an effective and accurate method to deduce the biological functions of novel sequences with high accuracy. As the use of experiments to validate the function of biological sequence is too expensive and hardly to be applied to large-scale data, the use of computer for prediction of gene function has become an economical and effective substitute. This paper proposes a new design of BLAST-based GO term annotator which incorporates data mining techniques and utilizes rough set theory. Moreover, this method is an evolution against the traditional methods which only base on BLAST or characters of GO Terms. Finally, experimental results prove the validity of the proposed rough set-based method.
Chapter PDF
Similar content being viewed by others
Keywords
References
Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H.J., Cherry, M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.J., Harris, M.A., Hill, D.P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J.C., Richardson, J.E., Ringwald, M., Rubin, G.M., Sherlock, G.: Gene Ontology: tool for the unification of biology. Nature Genetics 25, 25–29 (2000)
Pawlak, Z.: Rough Sets: Theoretical Aspects of Reasoning about Data. Kluwer, Dordrecht (1992)
Altschul, S., Gish, W., Miller, W., Myers, E., Lipman, D.: Basic Local Alignment Search Tool. Journal of Molecular Biology 215, 403–410 (1990)
Altschul, S.F., Madden, T.L., Schaffer, A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J.: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25, 3389–3402 (1997)
Hennig, S., Groth, D., Lehrach, H.: Automated Gene Ontology annotation for anonymous sequence data. Nucleic Acids Research 31, 3712–3715 (2003)
Groth, D., Lehrach, H., Hennig, S.: GOblet: a platform for Gene Ontology annotation of anonymous sequence data. Nucleic Acids Research 32, W313–W317 (2004)
Khan, S., Situ, G., Decker, K., Schmidt, C.J.: GoFigure: Automated Gene Ontology annotation. Bioinformatics 19, 2484–2485 (2003)
Martin, D.M.A., Berriman, M., Barton, G.J.: GOtcha: a new method for prediction of protein function assessed by the annotation of seven genomes. BMC Bioinformatics 5, 178 (2004)
Joslyn, C., Mniszewski, S., Fulmer, A., Heaton, G.: The Gene Ontology Categorizer. Bioinformatics 20, i169–i177 (2004)
Verspoor, K., Cohn, J., Mniszewski, S., Joslyn, C.: A Categorization Approach to Automated Ontological Protein Function Annotation. Protein Science 15, 1544–1549 (2006)
Author information
Authors and Affiliations
Editor information
Rights and permissions
Copyright information
© 2007 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Tang, XN., Lian, ZC., Pei, ZL., Liang, YC. (2007). Automated Methods of Predicting the Function of Biological Sequences Using GO and Rough Set. In: Rajapakse, J.C., Schmidt, B., Volkert, G. (eds) Pattern Recognition in Bioinformatics. PRIB 2007. Lecture Notes in Computer Science(), vol 4774. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-75286-8_1
Download citation
DOI: https://doi.org/10.1007/978-3-540-75286-8_1
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-75285-1
Online ISBN: 978-3-540-75286-8
eBook Packages: Computer ScienceComputer Science (R0)